Gene Page: DYNLL2
Summary ?
| GeneID | 140735 |
| Symbol | DYNLL2 |
| Synonyms | DNCL1B|Dlc2|RSPH22 |
| Description | dynein light chain LC8-type 2 |
| Reference | MIM:608942|HGNC:HGNC:24596|HPRD:06489| |
| Gene type | protein-coding |
| Map location | 17q22 |
| Pascal p-value | 0.57 |
| Sherlock p-value | 0.522 |
| Fetal beta | -0.936 |
| Support | INTRACELLULAR TRAFFICKING G2Cdb.humanPSD G2Cdb.humanPSP CompositeSet |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
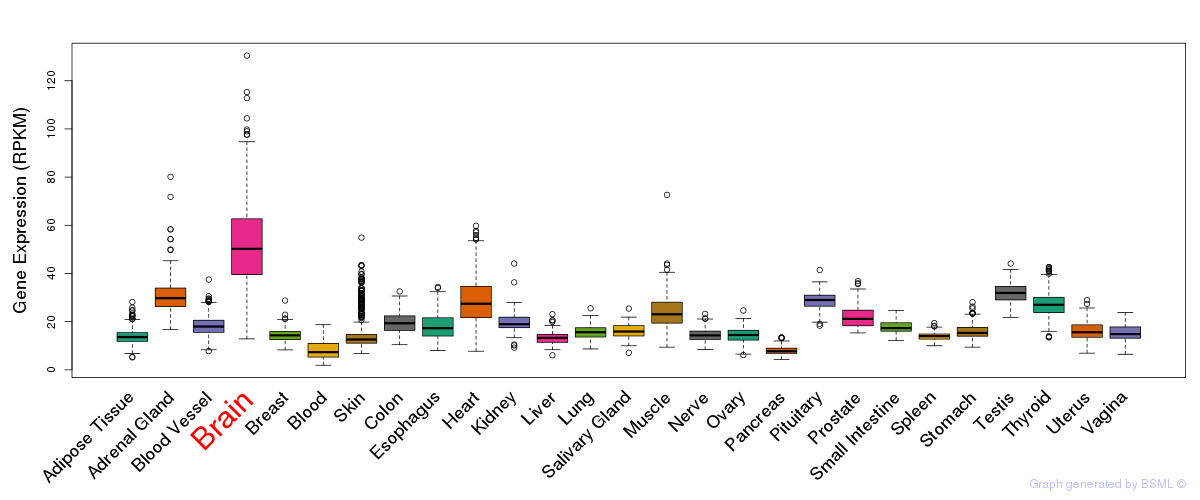
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| BCL2L11 | BAM | BIM | BIM-alpha6 | BIM-beta6 | BIM-beta7 | BOD | BimEL | BimL | BCL2-like 11 (apoptosis facilitator) | Reconstituted Complex | BioGRID | 14561217 |
| BMF | FLJ00065 | Bcl2 modifying factor | - | HPRD,BioGRID | 11546872 |14561217 |
| DLG2 | DKFZp781D1854 | DKFZp781E0954 | FLJ37266 | MGC131811 | PSD-93 | discs, large homolog 2, chapsyn-110 (Drosophila) | Affinity Capture-Western | BioGRID | 10844022 |
| DLG4 | FLJ97752 | FLJ98574 | PSD95 | SAP90 | discs, large homolog 4 (Drosophila) | - | HPRD,BioGRID | 10844022 |
| DLGAP1 | DAP-1 | DAP-1-ALPHA | DAP-1-BETA | GKAP | MGC88156 | SAPAP1 | hGKAP | discs, large (Drosophila) homolog-associated protein 1 | DAP-1 interacts with DLC-2. | BIND | 11122378 |
| DLGAP1 | DAP-1 | DAP-1-ALPHA | DAP-1-BETA | GKAP | MGC88156 | SAPAP1 | hGKAP | discs, large (Drosophila) homolog-associated protein 1 | - | HPRD,BioGRID | 10844022 |
| DNAL4 | PIG27 | dynein, axonemal, light chain 4 | Two-hybrid | BioGRID | 16189514 |
| DYNC1I1 | DNCI1 | DNCIC1 | dynein, cytoplasmic 1, intermediate chain 1 | Two-hybrid | BioGRID | 16189514 |
| DYNLL2 | DNCL1B | Dlc2 | MGC17810 | dynein, light chain, LC8-type 2 | Co-crystal Structure | BioGRID | 14561217 |
| EWSR1 | EWS | Ewing sarcoma breakpoint region 1 | Two-hybrid | BioGRID | 16189514 |
| GPHN | GEPH | GPH | GPHRYN | KIAA1385 | gephyrin | - | HPRD | 12097491 |
| GPRIN2 | GRIN2 | KIAA0514 | MGC15171 | G protein regulated inducer of neurite outgrowth 2 | Two-hybrid | BioGRID | 16189514 |
| HOMER3 | HOMER-3 | homer homolog 3 (Drosophila) | Two-hybrid | BioGRID | 16189514 |
| MYO5A | GS1 | MYH12 | MYO5 | MYR12 | myosin VA (heavy chain 12, myoxin) | Affinity Capture-Western Reconstituted Complex Two-hybrid | BioGRID | 10844022 |11546872 |
| SHANK2 | CORTBP1 | CTTNBP1 | ProSAP1 | SHANK | SPANK-3 | SH3 and multiple ankyrin repeat domains 2 | Affinity Capture-Western | BioGRID | 10844022 |
| THAP10 | - | THAP domain containing 10 | Two-hybrid | BioGRID | 16189514 |
| XIAP | API3 | BIRC4 | ILP1 | MIHA | XLP2 | X-linked inhibitor of apoptosis | Two-hybrid | BioGRID | 16189514 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG VASOPRESSIN REGULATED WATER REABSORPTION | 44 | 30 | All SZGR 2.0 genes in this pathway |
| REACTOME MHC CLASS II ANTIGEN PRESENTATION | 91 | 61 | All SZGR 2.0 genes in this pathway |
| REACTOME APOPTOSIS | 148 | 94 | All SZGR 2.0 genes in this pathway |
| REACTOME IMMUNE SYSTEM | 933 | 616 | All SZGR 2.0 genes in this pathway |
| REACTOME ACTIVATION OF BH3 ONLY PROTEINS | 17 | 12 | All SZGR 2.0 genes in this pathway |
| REACTOME ADAPTIVE IMMUNE SYSTEM | 539 | 350 | All SZGR 2.0 genes in this pathway |
| REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | 30 | 21 | All SZGR 2.0 genes in this pathway |
| WANG RESPONSE TO ANDROGEN UP | 29 | 21 | All SZGR 2.0 genes in this pathway |
| WANG RESPONSE TO FORSKOLIN UP | 23 | 17 | All SZGR 2.0 genes in this pathway |
| OSMAN BLADDER CANCER UP | 404 | 246 | All SZGR 2.0 genes in this pathway |
| MARKEY RB1 ACUTE LOF DN | 228 | 137 | All SZGR 2.0 genes in this pathway |
| MARTORIATI MDM4 TARGETS FETAL LIVER UP | 227 | 137 | All SZGR 2.0 genes in this pathway |
| NUYTTEN EZH2 TARGETS DN | 1024 | 594 | All SZGR 2.0 genes in this pathway |
| WEI MYCN TARGETS WITH E BOX | 795 | 478 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 6HR DN | 514 | 330 | All SZGR 2.0 genes in this pathway |
| DEN INTERACT WITH LCA5 | 26 | 21 | All SZGR 2.0 genes in this pathway |
| PASQUALUCCI LYMPHOMA BY GC STAGE UP | 283 | 177 | All SZGR 2.0 genes in this pathway |
| BYSTRYKH HEMATOPOIESIS STEM CELL QTL CIS | 128 | 77 | All SZGR 2.0 genes in this pathway |
| GOLDRATH HOMEOSTATIC PROLIFERATION | 171 | 102 | All SZGR 2.0 genes in this pathway |
| BYSTRYKH HEMATOPOIESIS STEM CELL AND BRAIN QTL CIS | 65 | 38 | All SZGR 2.0 genes in this pathway |
| CHESLER BRAIN HIGHEST EXPRESSION | 40 | 29 | All SZGR 2.0 genes in this pathway |
| ACEVEDO NORMAL TISSUE ADJACENT TO LIVER TUMOR DN | 354 | 216 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER CANCER DN | 540 | 340 | All SZGR 2.0 genes in this pathway |
| NAKAMURA METASTASIS MODEL UP | 45 | 26 | All SZGR 2.0 genes in this pathway |
| WONG ADULT TISSUE STEM MODULE | 721 | 492 | All SZGR 2.0 genes in this pathway |