Gene Page: CRYBB2
Summary ?
| GeneID | 1415 |
| Symbol | CRYBB2 |
| Synonyms | CCA2|CRYB2|CRYB2A|CTRCT3|D22S665 |
| Description | crystallin beta B2 |
| Reference | MIM:123620|HGNC:HGNC:2398|Ensembl:ENSG00000244752|HPRD:00430|Vega:OTTHUMG00000150905 |
| Gene type | protein-coding |
| Map location | 22q11.23 |
| Pascal p-value | 0.222 |
| Fetal beta | -0.214 |
| eGene | Myers' cis & trans Meta |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| GSMA_I | Genome scan meta-analysis | Psr: 0.031 | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs6004667 | chr22 | 25874463 | CRYBB2 | 1415 | 5.776E-37 | cis | ||
| rs6004668 | chr22 | 25874501 | CRYBB2 | 1415 | 1.157E-36 | cis | ||
| rs1930961 | chr22 | 25875264 | CRYBB2 | 1415 | 4.025E-33 | cis | ||
| rs6004673 | chr22 | 25885352 | CRYBB2 | 1415 | 3.915E-36 | cis | ||
| rs9612986 | chr22 | 25931436 | CRYBB2 | 1415 | 0.03 | cis | ||
| rs17103715 | chr1 | 48214622 | CRYBB2 | 1415 | 0.13 | trans | ||
| rs12087028 | chr1 | 214084383 | CRYBB2 | 1415 | 0.16 | trans | ||
| rs1005427 | chr2 | 39831229 | CRYBB2 | 1415 | 0.02 | trans | ||
| rs17030866 | chr2 | 66039792 | CRYBB2 | 1415 | 0 | trans | ||
| rs17006586 | chr2 | 71191207 | CRYBB2 | 1415 | 5.282E-4 | trans | ||
| rs17043357 | chr2 | 115445394 | CRYBB2 | 1415 | 0.05 | trans | ||
| rs10180903 | chr2 | 223115728 | CRYBB2 | 1415 | 0.15 | trans | ||
| rs11942948 | chr4 | 5254657 | CRYBB2 | 1415 | 0.07 | trans | ||
| rs10109794 | chr8 | 67045936 | CRYBB2 | 1415 | 0.08 | trans | ||
| rs1870392 | chr8 | 67098510 | CRYBB2 | 1415 | 0.08 | trans | ||
| rs17149833 | chr10 | 11247962 | CRYBB2 | 1415 | 0.06 | trans | ||
| rs11027761 | chr11 | 24217623 | CRYBB2 | 1415 | 0.11 | trans | ||
| rs17760954 | chr15 | 24258346 | CRYBB2 | 1415 | 0.01 | trans | ||
| rs17761129 | chr15 | 24262387 | CRYBB2 | 1415 | 0.1 | trans | ||
| rs6004667 | chr22 | 25874463 | CRYBB2 | 1415 | 1.788E-34 | trans | ||
| rs6004668 | chr22 | 25874501 | CRYBB2 | 1415 | 3.836E-34 | trans | ||
| rs1930961 | chr22 | 25875264 | CRYBB2 | 1415 | 1.363E-30 | trans | ||
| rs6004673 | chr22 | 25885352 | CRYBB2 | 1415 | 1.515E-33 | trans | ||
| rs9620540 | 0 | CRYBB2 | 1415 | 0 | trans | |||
| rs10521410 | chrX | 38116721 | CRYBB2 | 1415 | 0.06 | trans | ||
| rs5984472 | chrX | 89331417 | CRYBB2 | 1415 | 0.02 | trans | ||
| rs471777 | chrX | 96382529 | CRYBB2 | 1415 | 0.16 | trans | ||
| rs5966995 | chrX | 96387170 | CRYBB2 | 1415 | 0.16 | trans | ||
| rs10481828 | chrX | 115505090 | CRYBB2 | 1415 | 0.07 | trans | ||
| rs5952167 | chrX | 115529371 | CRYBB2 | 1415 | 0.01 | trans | ||
| rs12353619 | chrX | 118408162 | CRYBB2 | 1415 | 0.02 | trans | ||
| rs16999361 | chrX | 123792007 | CRYBB2 | 1415 | 0 | trans | ||
| rs453855 | chrX | 145778002 | CRYBB2 | 1415 | 0.07 | trans | ||
| rs6653376 | 0 | CRYBB2 | 1415 | 0.04 | trans |
Section II. Transcriptome annotation
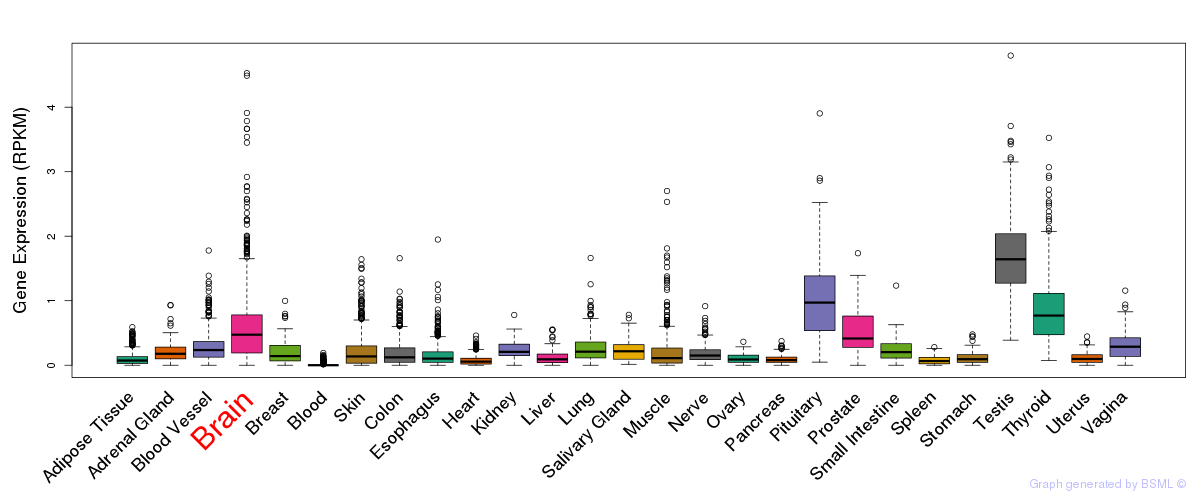
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005212 | structural constituent of eye lens | IEA | - | |
| GO:0042803 | protein homodimerization activity | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007601 | visual perception | TAS | 9158139 | |
| GO:0050896 | response to stimulus | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| PENG LEUCINE DEPRIVATION UP | 142 | 93 | All SZGR 2.0 genes in this pathway |
| KIM GASTRIC CANCER CHEMOSENSITIVITY | 103 | 64 | All SZGR 2.0 genes in this pathway |
| LY AGING MIDDLE UP | 14 | 11 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS UP | 1305 | 895 | All SZGR 2.0 genes in this pathway |
| HAN SATB1 TARGETS DN | 442 | 275 | All SZGR 2.0 genes in this pathway |
| GREGORY SYNTHETIC LETHAL WITH IMATINIB | 145 | 83 | All SZGR 2.0 genes in this pathway |
| LU EZH2 TARGETS UP | 295 | 155 | All SZGR 2.0 genes in this pathway |
| CERIBELLI GENES INACTIVE AND BOUND BY NFY | 45 | 27 | All SZGR 2.0 genes in this pathway |
| KRIEG HYPOXIA NOT VIA KDM3A | 770 | 480 | All SZGR 2.0 genes in this pathway |