Gene Page: LDLRAD3
Summary ?
| GeneID | 143458 |
| Symbol | LDLRAD3 |
| Synonyms | LRAD3 |
| Description | low density lipoprotein receptor class A domain containing 3 |
| Reference | HGNC:HGNC:27046|Ensembl:ENSG00000179241|HPRD:14063|Vega:OTTHUMG00000166313 |
| Gene type | protein-coding |
| Map location | 11p13 |
| Pascal p-value | 0.335 |
| TADA p-value | 0.01 |
| Fetal beta | 0.886 |
| eGene | Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DNM:McCarthy_2014 | Whole Exome Sequencing analysis | Whole exome sequencing of 57 trios with sporadic or familial schizophrenia. |
Section I. Genetics and epigenetics annotation
 DNM table
DNM table
| Gene | Chromosome | Position | Ref | Alt | Transcript | AA change | Mutation type | Sift | CG46 | Trait | Study |
|---|---|---|---|---|---|---|---|---|---|---|---|
| LDLRAD3 | chr11 | 36248656 | T | C | NM_174902 | p.F159S | missense SNV | C | P | Schizophrenia | DNM:McCarthy_2014 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs2201845 | chr11 | 36556257 | LDLRAD3 | 143458 | 0.14 | cis | ||
| rs6753473 | chr2 | 26526418 | LDLRAD3 | 143458 | 0.05 | trans | ||
| rs16829545 | chr2 | 151977407 | LDLRAD3 | 143458 | 4.466E-13 | trans | ||
| rs7584986 | chr2 | 184111432 | LDLRAD3 | 143458 | 0.03 | trans | ||
| rs16860529 | chr3 | 185900368 | LDLRAD3 | 143458 | 0.05 | trans | ||
| rs10035168 | chr5 | 5974295 | LDLRAD3 | 143458 | 0.2 | trans | ||
| rs10474151 | chr5 | 84286402 | LDLRAD3 | 143458 | 0.1 | trans | ||
| rs13360496 | 0 | LDLRAD3 | 143458 | 2.194E-4 | trans | |||
| rs12196880 | chr6 | 24313746 | LDLRAD3 | 143458 | 0.18 | trans | ||
| rs16890367 | chr6 | 38078448 | LDLRAD3 | 143458 | 0.2 | trans | ||
| rs7787830 | chr7 | 98797019 | LDLRAD3 | 143458 | 0.06 | trans | ||
| rs11139334 | chr9 | 84209393 | LDLRAD3 | 143458 | 0.07 | trans | ||
| rs3765543 | chr9 | 134458323 | LDLRAD3 | 143458 | 0.02 | trans | ||
| rs2393316 | chr10 | 59333070 | LDLRAD3 | 143458 | 0.02 | trans | ||
| rs17124813 | chr10 | 110347464 | LDLRAD3 | 143458 | 0.17 | trans | ||
| rs4112777 | chr13 | 99281457 | LDLRAD3 | 143458 | 0.13 | trans | ||
| rs1279323 | chr14 | 33585158 | LDLRAD3 | 143458 | 0.01 | trans | ||
| rs9989228 | chr14 | 37809252 | LDLRAD3 | 143458 | 0.06 | trans | ||
| rs16955618 | chr15 | 29937543 | LDLRAD3 | 143458 | 5.843E-23 | trans | ||
| rs2077735 | chr15 | 58479024 | LDLRAD3 | 143458 | 0.14 | trans | ||
| rs1041786 | chr21 | 22617710 | LDLRAD3 | 143458 | 0.04 | trans | ||
| rs3829726 | chrX | 72666188 | LDLRAD3 | 143458 | 0.2 | trans |
Section II. Transcriptome annotation
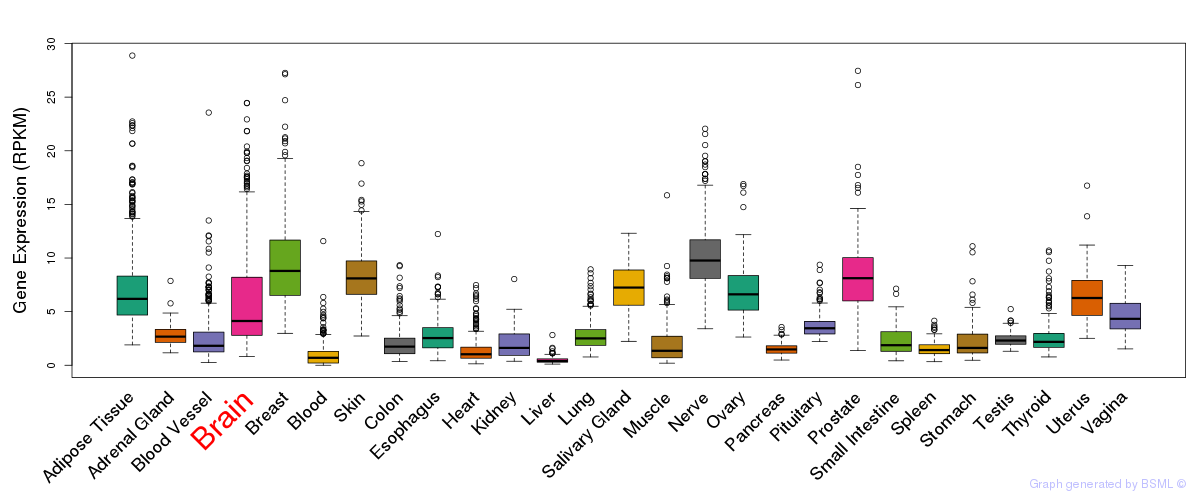
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| FULCHER INFLAMMATORY RESPONSE LECTIN VS LPS DN | 463 | 290 | All SZGR 2.0 genes in this pathway |
| OSMAN BLADDER CANCER UP | 404 | 246 | All SZGR 2.0 genes in this pathway |
| WEI MYCN TARGETS WITH E BOX | 795 | 478 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS EARLY PROGENITOR | 532 | 309 | All SZGR 2.0 genes in this pathway |
| RODWELL AGING KIDNEY UP | 487 | 303 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 6HR DN | 911 | 527 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 24HR DN | 1011 | 592 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 1 | 528 | 324 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 UNSTIMULATED | 1229 | 713 | All SZGR 2.0 genes in this pathway |
| RAY TUMORIGENESIS BY ERBB2 CDC25A DN | 159 | 105 | All SZGR 2.0 genes in this pathway |
| BREDEMEYER RAG SIGNALING VIA ATM NOT VIA NFKB UP | 49 | 32 | All SZGR 2.0 genes in this pathway |
| VANASSE BCL2 TARGETS DN | 74 | 50 | All SZGR 2.0 genes in this pathway |
| CHIANG LIVER CANCER SUBCLASS PROLIFERATION UP | 178 | 108 | All SZGR 2.0 genes in this pathway |
| DUTERTRE ESTRADIOL RESPONSE 6HR UP | 229 | 149 | All SZGR 2.0 genes in this pathway |
| CHYLA CBFA2T3 TARGETS UP | 387 | 225 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS DN | 1972 | 1213 | All SZGR 2.0 genes in this pathway |
| GOBERT OLIGODENDROCYTE DIFFERENTIATION DN | 1080 | 713 | All SZGR 2.0 genes in this pathway |
| WAKABAYASHI ADIPOGENESIS PPARG RXRA BOUND 8D | 882 | 506 | All SZGR 2.0 genes in this pathway |
| KRIEG KDM3A TARGETS NOT HYPOXIA | 208 | 107 | All SZGR 2.0 genes in this pathway |