Gene Page: CSF2RB
Summary ?
| GeneID | 1439 |
| Symbol | CSF2RB |
| Synonyms | CD131|CDw131|IL3RB|IL5RB|SMDP5 |
| Description | colony stimulating factor 2 receptor beta common subunit |
| Reference | MIM:138981|HGNC:HGNC:2436|Ensembl:ENSG00000100368|HPRD:00739|Vega:OTTHUMG00000150546 |
| Gene type | protein-coding |
| Map location | 22q13.1 |
| Pascal p-value | 0.09 |
| Fetal beta | -0.125 |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Association | A combined odds ratio method (Sun et al. 2008), association studies | 2 | Link to SZGene |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.0294 |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
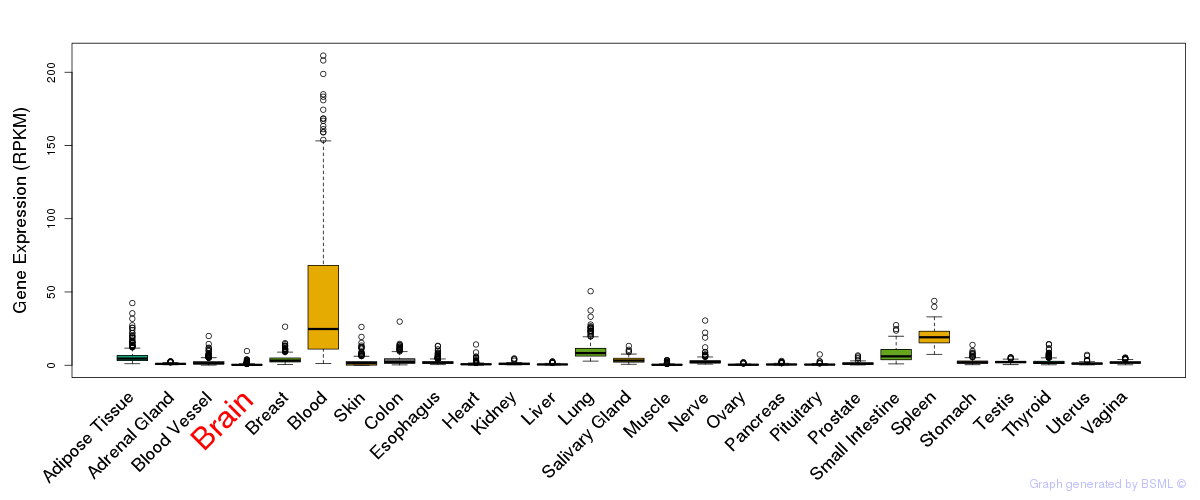
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0004872 | receptor activity | TAS | 9410898 | |
| GO:0004914 | interleukin-5 receptor activity | TAS | 9410898 | |
| GO:0004912 | interleukin-3 receptor activity | TAS | 9410898 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007165 | signal transduction | NAS | 1702217 | |
| GO:0007585 | respiratory gaseous exchange | TAS | 9410898 | |
| GO:0019221 | cytokine-mediated signaling pathway | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0016020 | membrane | IEA | - | |
| GO:0030526 | granulocyte macrophage colony-stimulating factor receptor complex | TAS | 9410898 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| CSF2 | GMCSF | MGC131935 | MGC138897 | colony stimulating factor 2 (granulocyte-macrophage) | - | HPRD | 2645577 |8278375 |
| CSF2RA | CD116 | CDw116 | CSF2R | CSF2RAX | CSF2RAY | CSF2RX | CSF2RY | GM-CSF-R-alpha | GMCSFR | GMR | MGC3848 | MGC4838 | colony stimulating factor 2 receptor, alpha, low-affinity (granulocyte-macrophage) | - | HPRD | 9211889 |
| CSF2RB | CD131 | CDw131 | IL3RB | IL5RB | colony stimulating factor 2 receptor, beta, low-affinity (granulocyte-macrophage) | - | HPRD,BioGRID | 11207369 |
| EPOR | MGC138358 | erythropoietin receptor | - | HPRD | 12488507 |
| GNB2L1 | Gnb2-rs1 | H12.3 | HLC-7 | PIG21 | RACK1 | guanine nucleotide binding protein (G protein), beta polypeptide 2-like 1 | - | HPRD,BioGRID | 10490850 |
| IL3 | IL-3 | MCGF | MGC79398 | MGC79399 | MULTI-CSF | interleukin 3 (colony-stimulating factor, multiple) | Reconstituted Complex | BioGRID | 8649415 |
| IL3 | IL-3 | MCGF | MGC79398 | MGC79399 | MULTI-CSF | interleukin 3 (colony-stimulating factor, multiple) | - | HPRD | 9278420 |
| IL5RA | CD125 | CDw125 | HSIL5R3 | IL5R | MGC26560 | interleukin 5 receptor, alpha | Affinity Capture-Western | BioGRID | 9516124 |
| JAK2 | JTK10 | Janus kinase 2 (a protein tyrosine kinase) | Affinity Capture-Western in vivo Reconstituted Complex | BioGRID | 7527392 |7775438 |10772872 |
| LYN | FLJ26625 | JTK8 | v-yes-1 Yamaguchi sarcoma viral related oncogene homolog | - | HPRD | 7636265|9973406 |
| LYN | FLJ26625 | JTK8 | v-yes-1 Yamaguchi sarcoma viral related oncogene homolog | in vivo Protein-peptide | BioGRID | 7636265 |9973406 |
| MAD2L1 | HSMAD2 | MAD2 | MAD2 mitotic arrest deficient-like 1 (yeast) | - | HPRD,BioGRID | 11551900 |
| MAD2L1 | HSMAD2 | MAD2 | MAD2 mitotic arrest deficient-like 1 (yeast) | MAD2 interacts with CSF2RB. This interaction was modelled on a demonstrated interaction between mouse MAD2 and human CSFR2B. | BIND | 11551900 |
| PRKCB | MGC41878 | PKC-beta | PKCB | PRKCB1 | PRKCB2 | protein kinase C, beta | Affinity Capture-Western | BioGRID | 10490850 |
| PTPN11 | BPTP3 | CFC | MGC14433 | NS1 | PTP-1D | PTP2C | SH-PTP2 | SH-PTP3 | SHP2 | protein tyrosine phosphatase, non-receptor type 11 | - | HPRD,BioGRID | 9162089 |
| PTPN6 | HCP | HCPH | HPTP1C | PTP-1C | SH-PTP1 | SHP-1 | SHP-1L | SHP1 | protein tyrosine phosphatase, non-receptor type 6 | - | HPRD,BioGRID | 8246974 |
| SHC1 | FLJ26504 | SHC | SHCA | SHC (Src homology 2 domain containing) transforming protein 1 | - | HPRD,BioGRID | 10477722 |10704825 |
| YWHAZ | KCIP-1 | MGC111427 | MGC126532 | MGC138156 | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide | - | HPRD,BioGRID | 8278375 |10477722 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG CYTOKINE CYTOKINE RECEPTOR INTERACTION | 267 | 161 | All SZGR 2.0 genes in this pathway |
| KEGG APOPTOSIS | 88 | 62 | All SZGR 2.0 genes in this pathway |
| KEGG JAK STAT SIGNALING PATHWAY | 155 | 105 | All SZGR 2.0 genes in this pathway |
| BIOCARTA IL3 PATHWAY | 15 | 11 | All SZGR 2.0 genes in this pathway |
| BIOCARTA BAD PATHWAY | 26 | 23 | All SZGR 2.0 genes in this pathway |
| PID GMCSF PATHWAY | 37 | 31 | All SZGR 2.0 genes in this pathway |
| PID IL5 PATHWAY | 14 | 11 | All SZGR 2.0 genes in this pathway |
| PID IL3 PATHWAY | 27 | 19 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY ILS | 107 | 86 | All SZGR 2.0 genes in this pathway |
| REACTOME IL 3 5 AND GM CSF SIGNALING | 43 | 34 | All SZGR 2.0 genes in this pathway |
| REACTOME IL RECEPTOR SHC SIGNALING | 27 | 21 | All SZGR 2.0 genes in this pathway |
| REACTOME IL 2 SIGNALING | 41 | 34 | All SZGR 2.0 genes in this pathway |
| REACTOME IMMUNE SYSTEM | 933 | 616 | All SZGR 2.0 genes in this pathway |
| REACTOME CYTOKINE SIGNALING IN IMMUNE SYSTEM | 270 | 204 | All SZGR 2.0 genes in this pathway |
| SCHUETZ BREAST CANCER DUCTAL INVASIVE UP | 351 | 230 | All SZGR 2.0 genes in this pathway |
| CASORELLI ACUTE PROMYELOCYTIC LEUKEMIA DN | 663 | 425 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP | 1382 | 904 | All SZGR 2.0 genes in this pathway |
| TONKS TARGETS OF RUNX1 RUNX1T1 FUSION HSC DN | 187 | 115 | All SZGR 2.0 genes in this pathway |
| PAPASPYRIDONOS UNSTABLE ATEROSCLEROTIC PLAQUE UP | 52 | 36 | All SZGR 2.0 genes in this pathway |
| GRAHAM NORMAL QUIESCENT VS NORMAL DIVIDING DN | 87 | 49 | All SZGR 2.0 genes in this pathway |
| WANG ESOPHAGUS CANCER VS NORMAL UP | 121 | 72 | All SZGR 2.0 genes in this pathway |
| MARTORIATI MDM4 TARGETS NEUROEPITHELIUM DN | 164 | 111 | All SZGR 2.0 genes in this pathway |
| MARTORIATI MDM4 TARGETS FETAL LIVER DN | 514 | 319 | All SZGR 2.0 genes in this pathway |
| JAZAG TGFB1 SIGNALING UP | 108 | 69 | All SZGR 2.0 genes in this pathway |
| PUJANA ATM PCC NETWORK | 1442 | 892 | All SZGR 2.0 genes in this pathway |
| BENPORATH MYC MAX TARGETS | 775 | 494 | All SZGR 2.0 genes in this pathway |
| GALINDO IMMUNE RESPONSE TO ENTEROTOXIN | 85 | 67 | All SZGR 2.0 genes in this pathway |
| ZHAN MULTIPLE MYELOMA CD1 AND CD2 DN | 51 | 32 | All SZGR 2.0 genes in this pathway |
| ZHAN MULTIPLE MYELOMA LB UP | 48 | 28 | All SZGR 2.0 genes in this pathway |
| BASSO CD40 SIGNALING DN | 68 | 44 | All SZGR 2.0 genes in this pathway |
| YAGI AML FAB MARKERS | 191 | 131 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS LATE PROGENITOR | 544 | 307 | All SZGR 2.0 genes in this pathway |
| MCLACHLAN DENTAL CARIES DN | 245 | 144 | All SZGR 2.0 genes in this pathway |
| MCLACHLAN DENTAL CARIES UP | 253 | 147 | All SZGR 2.0 genes in this pathway |
| KYNG DNA DAMAGE BY UV | 62 | 43 | All SZGR 2.0 genes in this pathway |
| KYNG DNA DAMAGE DN | 195 | 135 | All SZGR 2.0 genes in this pathway |
| STEARMAN TUMOR FIELD EFFECT UP | 36 | 22 | All SZGR 2.0 genes in this pathway |
| STEARMAN LUNG CANCER EARLY VS LATE DN | 61 | 39 | All SZGR 2.0 genes in this pathway |
| HELLER SILENCED BY METHYLATION UP | 282 | 183 | All SZGR 2.0 genes in this pathway |
| WALLACE PROSTATE CANCER RACE UP | 299 | 167 | All SZGR 2.0 genes in this pathway |
| MARZEC IL2 SIGNALING UP | 115 | 80 | All SZGR 2.0 genes in this pathway |
| MCCABE BOUND BY HOXC6 | 469 | 239 | All SZGR 2.0 genes in this pathway |
| BOQUEST STEM CELL DN | 216 | 143 | All SZGR 2.0 genes in this pathway |
| WANG TUMOR INVASIVENESS DN | 210 | 128 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS UP | 1305 | 895 | All SZGR 2.0 genes in this pathway |
| POOLA INVASIVE BREAST CANCER UP | 288 | 168 | All SZGR 2.0 genes in this pathway |
| DANG BOUND BY MYC | 1103 | 714 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 1HR DN | 222 | 147 | All SZGR 2.0 genes in this pathway |
| MARTENS TRETINOIN RESPONSE UP | 857 | 456 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS DN | 1972 | 1213 | All SZGR 2.0 genes in this pathway |
| BAKKER FOXO3 TARGETS UP | 61 | 41 | All SZGR 2.0 genes in this pathway |
| ALTEMEIER RESPONSE TO LPS WITH MECHANICAL VENTILATION | 128 | 81 | All SZGR 2.0 genes in this pathway |