Gene Page: CSNK1A1
Summary ?
| GeneID | 1452 |
| Symbol | CSNK1A1 |
| Synonyms | CK1|CK1a|CKIa|HEL-S-77p|HLCDGP1|PRO2975 |
| Description | casein kinase 1 alpha 1 |
| Reference | MIM:600505|HGNC:HGNC:2451|Ensembl:ENSG00000113712|HPRD:02739|Vega:OTTHUMG00000163463 |
| Gene type | protein-coding |
| Map location | 5q32 |
| Pascal p-value | 0.214 |
| Sherlock p-value | 0.843 |
| Fetal beta | 1.434 |
| eGene | Myers' cis & trans |
| Support | G2Cdb.humanPSD G2Cdb.humanPSP |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| GSMA_I | Genome scan meta-analysis | Psr: 0.0032 | |
| GSMA_IIE | Genome scan meta-analysis (European-ancestry samples) | Psr: 0.01718 | |
| GSMA_IIA | Genome scan meta-analysis (All samples) | Psr: 0.00459 | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.0752 |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs4491879 | chr3 | 189373908 | CSNK1A1 | 1452 | 0.15 | trans |
Section II. Transcriptome annotation
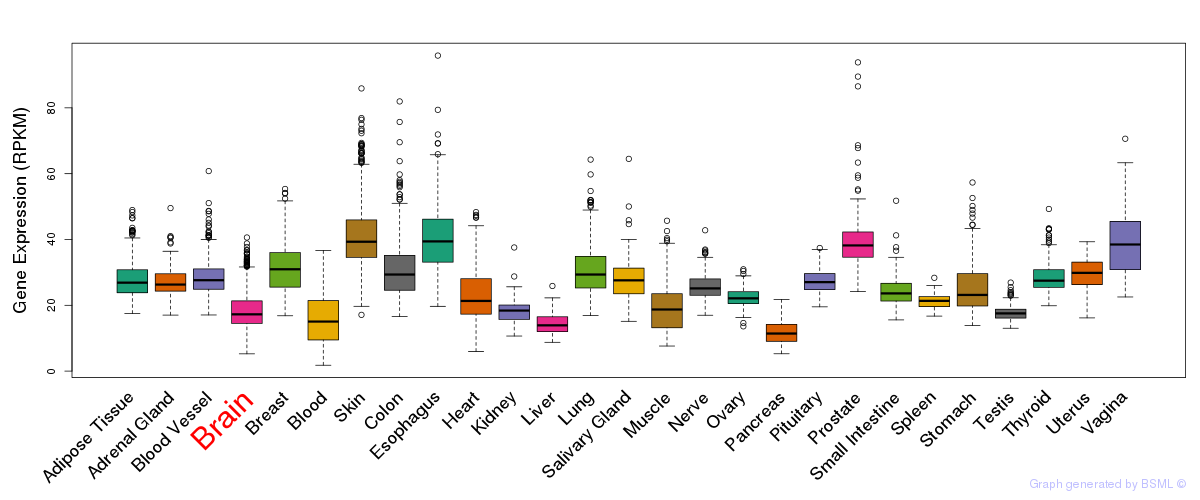
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| SEC61A1 | 0.91 | 0.90 |
| MTMR3 | 0.90 | 0.89 |
| ZXDC | 0.90 | 0.87 |
| SEPN1 | 0.89 | 0.92 |
| SUFU | 0.89 | 0.87 |
| PHF2 | 0.89 | 0.91 |
| AMMECR1L | 0.89 | 0.86 |
| LIX1L | 0.89 | 0.90 |
| POFUT1 | 0.89 | 0.86 |
| PATZ1 | 0.89 | 0.90 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| C5orf53 | -0.71 | -0.76 |
| AF347015.27 | -0.66 | -0.76 |
| S100B | -0.65 | -0.74 |
| ACOT13 | -0.64 | -0.68 |
| MT-CO2 | -0.64 | -0.78 |
| AF347015.31 | -0.64 | -0.76 |
| HLA-F | -0.64 | -0.66 |
| AF347015.33 | -0.63 | -0.74 |
| CA4 | -0.63 | -0.67 |
| FXYD1 | -0.63 | -0.73 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0000166 | nucleotide binding | IEA | - | |
| GO:0005524 | ATP binding | IEA | - | |
| GO:0004674 | protein serine/threonine kinase activity | IEA | - | |
| GO:0016740 | transferase activity | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006468 | protein amino acid phosphorylation | TAS | 10777483 | |
| GO:0016055 | Wnt receptor signaling pathway | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005829 | cytosol | EXP | 11955436 |12000790 |12820959 |15327769 |16753179 | |
| GO:0005737 | cytoplasm | IEA | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ACTA1 | ACTA | ASMA | CFTD | CFTD1 | CFTDM | MPFD | NEM1 | NEM2 | NEM3 | actin, alpha 1, skeletal muscle | Affinity Capture-MS | BioGRID | 12062430 |
| ADAP1 | CENTA1 | GCS1L | p42IP4 | ArfGAP with dual PH domains 1 | - | HPRD,BioGRID | 11278595 |
| AP3B1 | ADTB3 | ADTB3A | HPS | HPS2 | PE | adaptor-related protein complex 3, beta 1 subunit | - | HPRD | 12062430 |
| AXIN1 | AXIN | MGC52315 | axin 1 | - | HPRD,BioGRID | 11884395 |
| CHRM3 | HM3 | cholinergic receptor, muscarinic 3 | - | HPRD,BioGRID | 10777483 |
| ERF | PE-2 | PE2 | Ets2 repressor factor | Co-purification | BioGRID | 12062430 |
| HMGB1 | DKFZp686A04236 | HMG1 | HMG3 | SBP-1 | high-mobility group box 1 | - | HPRD,BioGRID | 12062430 |
| HMGB2 | HMG2 | high-mobility group box 2 | - | HPRD,BioGRID | 12062430 |
| KPNA2 | IPOA1 | QIP2 | RCH1 | SRP1alpha | karyopherin alpha 2 (RAG cohort 1, importin alpha 1) | - | HPRD,BioGRID | 12062430 |
| PPP1R14A | CPI-17 | CPI17 | PPP1INL | protein phosphatase 1, regulatory (inhibitor) subunit 14A | - | HPRD,BioGRID | 12062430 |
| PPP1R14A | CPI-17 | CPI17 | PPP1INL | protein phosphatase 1, regulatory (inhibitor) subunit 14A | - | HPRD | 15003508 |
| PPP2CA | PP2Ac | PP2CA | RP-C | protein phosphatase 2 (formerly 2A), catalytic subunit, alpha isoform | Affinity Capture-MS | BioGRID | 12062430 |
| PPP2R5A | B56A | MGC131915 | PR61A | protein phosphatase 2, regulatory subunit B', alpha isoform | - | HPRD | 12062430 |
| RCC1 | CHC1 | RCC1-I | regulator of chromosome condensation 1 | - | HPRD,BioGRID | 12062430 |
| SYT9 | FLJ45896 | synaptotagmin IX | - | HPRD,BioGRID | 12062430 |
| TNFRSF1B | CD120b | TBPII | TNF-R-II | TNF-R75 | TNFBR | TNFR1B | TNFR2 | TNFR80 | p75 | p75TNFR | tumor necrosis factor receptor superfamily, member 1B | - | HPRD,BioGRID | 7559483 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG WNT SIGNALING PATHWAY | 151 | 112 | All SZGR 2.0 genes in this pathway |
| KEGG HEDGEHOG SIGNALING PATHWAY | 56 | 42 | All SZGR 2.0 genes in this pathway |
| BIOCARTA P35ALZHEIMERS PATHWAY | 11 | 9 | All SZGR 2.0 genes in this pathway |
| BIOCARTA P53HYPOXIA PATHWAY | 23 | 21 | All SZGR 2.0 genes in this pathway |
| BIOCARTA NFAT PATHWAY | 56 | 45 | All SZGR 2.0 genes in this pathway |
| BIOCARTA WNT PATHWAY | 26 | 24 | All SZGR 2.0 genes in this pathway |
| WNT SIGNALING | 89 | 71 | All SZGR 2.0 genes in this pathway |
| ST WNT BETA CATENIN PATHWAY | 34 | 28 | All SZGR 2.0 genes in this pathway |
| ST FAS SIGNALING PATHWAY | 65 | 54 | All SZGR 2.0 genes in this pathway |
| PID WNT NONCANONICAL PATHWAY | 32 | 26 | All SZGR 2.0 genes in this pathway |
| PID BETA CATENIN DEG PATHWAY | 18 | 17 | All SZGR 2.0 genes in this pathway |
| PID PS1 PATHWAY | 46 | 39 | All SZGR 2.0 genes in this pathway |
| PID NFAT 3PATHWAY | 54 | 47 | All SZGR 2.0 genes in this pathway |
| PID FOXO PATHWAY | 49 | 43 | All SZGR 2.0 genes in this pathway |
| PID HEDGEHOG GLI PATHWAY | 48 | 35 | All SZGR 2.0 genes in this pathway |
| PID P53 REGULATION PATHWAY | 59 | 50 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY WNT | 65 | 41 | All SZGR 2.0 genes in this pathway |
| REACTOME CTNNB1 PHOSPHORYLATION CASCADE | 16 | 11 | All SZGR 2.0 genes in this pathway |
| PARENT MTOR SIGNALING UP | 567 | 375 | All SZGR 2.0 genes in this pathway |
| TURASHVILI BREAST DUCTAL CARCINOMA VS DUCTAL NORMAL DN | 198 | 110 | All SZGR 2.0 genes in this pathway |
| TURASHVILI BREAST DUCTAL CARCINOMA VS LOBULAR NORMAL DN | 69 | 43 | All SZGR 2.0 genes in this pathway |
| THUM SYSTOLIC HEART FAILURE DN | 244 | 147 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP | 1382 | 904 | All SZGR 2.0 genes in this pathway |
| LAIHO COLORECTAL CANCER SERRATED UP | 112 | 71 | All SZGR 2.0 genes in this pathway |
| WANG LMO4 TARGETS UP | 372 | 227 | All SZGR 2.0 genes in this pathway |
| OSMAN BLADDER CANCER UP | 404 | 246 | All SZGR 2.0 genes in this pathway |
| GINESTIER BREAST CANCER ZNF217 AMPLIFIED UP | 78 | 48 | All SZGR 2.0 genes in this pathway |
| TIEN INTESTINE PROBIOTICS 24HR DN | 214 | 133 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA POORLY DIFFERENTIATED UP | 633 | 376 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA ANAPLASTIC UP | 722 | 443 | All SZGR 2.0 genes in this pathway |
| HAHTOLA MYCOSIS FUNGOIDES SKIN DN | 27 | 20 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE KERATINOCYTE DN | 485 | 334 | All SZGR 2.0 genes in this pathway |
| GAUSSMANN MLL AF4 FUSION TARGETS C UP | 170 | 114 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 DN | 855 | 609 | All SZGR 2.0 genes in this pathway |
| GRUETZMANN PANCREATIC CANCER UP | 358 | 245 | All SZGR 2.0 genes in this pathway |
| SPIELMAN LYMPHOBLAST EUROPEAN VS ASIAN DN | 584 | 395 | All SZGR 2.0 genes in this pathway |
| RICKMAN TUMOR DIFFERENTIATED WELL VS POORLY DN | 382 | 224 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 6HR DN | 514 | 330 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT AND CANCER BOX4 DN | 32 | 22 | All SZGR 2.0 genes in this pathway |
| COLIN PILOCYTIC ASTROCYTOMA VS GLIOBLASTOMA DN | 28 | 21 | All SZGR 2.0 genes in this pathway |
| STARK PREFRONTAL CORTEX 22Q11 DELETION UP | 195 | 138 | All SZGR 2.0 genes in this pathway |
| GALE APL WITH FLT3 MUTATED UP | 56 | 35 | All SZGR 2.0 genes in this pathway |
| KUMAR TARGETS OF MLL AF9 FUSION | 405 | 264 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| WELCSH BRCA1 TARGETS UP | 198 | 132 | All SZGR 2.0 genes in this pathway |
| CARDOSO RESPONSE TO GAMMA RADIATION AND 3AB | 18 | 11 | All SZGR 2.0 genes in this pathway |
| JIANG AGING CEREBRAL CORTEX DN | 54 | 43 | All SZGR 2.0 genes in this pathway |
| BILD SRC ONCOGENIC SIGNATURE | 62 | 38 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 6HR UP | 953 | 554 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 24HR UP | 783 | 442 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 UNSTIMULATED | 1229 | 713 | All SZGR 2.0 genes in this pathway |
| KYNG DNA DAMAGE BY GAMMA AND UV RADIATION | 88 | 65 | All SZGR 2.0 genes in this pathway |
| KYNG DNA DAMAGE DN | 195 | 135 | All SZGR 2.0 genes in this pathway |
| FINETTI BREAST CANCERS KINOME GRAY | 15 | 10 | All SZGR 2.0 genes in this pathway |
| IWANAGA CARCINOGENESIS BY KRAS UP | 170 | 107 | All SZGR 2.0 genes in this pathway |
| BLUM RESPONSE TO SALIRASIB UP | 245 | 159 | All SZGR 2.0 genes in this pathway |
| VANTVEER BREAST CANCER BRCA1 DN | 44 | 28 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS UP | 504 | 321 | All SZGR 2.0 genes in this pathway |
| JOHNSTONE PARVB TARGETS 2 DN | 336 | 211 | All SZGR 2.0 genes in this pathway |
| JOHNSTONE PARVB TARGETS 3 DN | 918 | 550 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-135 | 958 | 964 | m8 | hsa-miR-135a | UAUGGCUUUUUAUUCCUAUGUGA |
| hsa-miR-135b | UAUGGCUUUUCAUUCCUAUGUG | ||||
| miR-141/200a | 550 | 556 | m8 | hsa-miR-141 | UAACACUGUCUGGUAAAGAUGG |
| hsa-miR-200a | UAACACUGUCUGGUAACGAUGU | ||||
| miR-155 | 662 | 668 | 1A | hsa-miR-155 | UUAAUGCUAAUCGUGAUAGGGG |
| miR-186 | 1073 | 1079 | m8 | hsa-miR-186 | CAAAGAAUUCUCCUUUUGGGCUU |
| miR-203.1 | 369 | 375 | 1A | hsa-miR-203 | UGAAAUGUUUAGGACCACUAG |
| miR-30-5p | 830 | 837 | 1A,m8 | hsa-miR-30a-5p | UGUAAACAUCCUCGACUGGAAG |
| hsa-miR-30cbrain | UGUAAACAUCCUACACUCUCAGC | ||||
| hsa-miR-30dSZ | UGUAAACAUCCCCGACUGGAAG | ||||
| hsa-miR-30bSZ | UGUAAACAUCCUACACUCAGCU | ||||
| hsa-miR-30e-5p | UGUAAACAUCCUUGACUGGA | ||||
| miR-325 | 638 | 645 | 1A,m8 | hsa-miR-325 | CCUAGUAGGUGUCCAGUAAGUGU |
| miR-330 | 356 | 363 | 1A,m8 | hsa-miR-330brain | GCAAAGCACACGGCCUGCAGAGA |
| miR-409-3p | 486 | 492 | m8 | hsa-miR-409-3p | CGAAUGUUGCUCGGUGAACCCCU |
| miR-493-5p | 83 | 89 | m8 | hsa-miR-493-5p | UUGUACAUGGUAGGCUUUCAUU |
| miR-495 | 326 | 332 | 1A | hsa-miR-495brain | AAACAAACAUGGUGCACUUCUUU |
| miR-9 | 539 | 546 | 1A,m8 | hsa-miR-9SZ | UCUUUGGUUAUCUAGCUGUAUGA |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.