Gene Page: CSRP2
Summary ?
| GeneID | 1466 |
| Symbol | CSRP2 |
| Synonyms | CRP2|LMO5|SmLIM |
| Description | cysteine and glycine rich protein 2 |
| Reference | MIM:601871|HGNC:HGNC:2470|Ensembl:ENSG00000175183|HPRD:03523|Vega:OTTHUMG00000169919 |
| Gene type | protein-coding |
| Map location | 12q21.1 |
| Pascal p-value | 0.493 |
| Sherlock p-value | 0.737 |
| Fetal beta | 4.013 |
| DMG | 1 (# studies) |
| eGene | Caudate basal ganglia Nucleus accumbens basal ganglia Putamen basal ganglia Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg10582318 | 12 | 77273011 | CSRP2 | 1.929E-4 | -0.352 | 0.035 | DMG:Wockner_2014 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs1868354 | chr12 | 77440580 | CSRP2 | 1466 | 0.18 | cis | ||
| rs1257641 | chr14 | 99480394 | CSRP2 | 1466 | 0.19 | trans | ||
| rs9612557 | chr22 | 24535629 | CSRP2 | 1466 | 0.07 | trans | ||
| rs1495333 | 12 | 77300447 | CSRP2 | ENSG00000175183.5 | 1.195E-8 | 0 | -27607 | gtex_brain_putamen_basal |
| rs201910500 | 12 | 77300644 | CSRP2 | ENSG00000175183.5 | 2.819E-9 | 0 | -27804 | gtex_brain_putamen_basal |
| rs773464 | 12 | 77304373 | CSRP2 | ENSG00000175183.5 | 4.706E-7 | 0 | -31533 | gtex_brain_putamen_basal |
| rs773463 | 12 | 77304825 | CSRP2 | ENSG00000175183.5 | 9.457E-8 | 0 | -31985 | gtex_brain_putamen_basal |
| rs398020216 | 12 | 77308842 | CSRP2 | ENSG00000175183.5 | 5.252E-10 | 0 | -36002 | gtex_brain_putamen_basal |
| rs7960465 | 12 | 77309358 | CSRP2 | ENSG00000175183.5 | 1.195E-8 | 0 | -36518 | gtex_brain_putamen_basal |
| rs7976747 | 12 | 77313694 | CSRP2 | ENSG00000175183.5 | 1.516E-9 | 0 | -40854 | gtex_brain_putamen_basal |
| rs12812426 | 12 | 77314197 | CSRP2 | ENSG00000175183.5 | 1.288E-9 | 0 | -41357 | gtex_brain_putamen_basal |
| rs1873913 | 12 | 77315273 | CSRP2 | ENSG00000175183.5 | 9.059E-10 | 0 | -42433 | gtex_brain_putamen_basal |
| rs11116211 | 12 | 77315824 | CSRP2 | ENSG00000175183.5 | 7.434E-10 | 0 | -42984 | gtex_brain_putamen_basal |
| rs773481 | 12 | 77318084 | CSRP2 | ENSG00000175183.5 | 4.973E-8 | 0 | -45244 | gtex_brain_putamen_basal |
| rs773484 | 12 | 77319533 | CSRP2 | ENSG00000175183.5 | 5.295E-7 | 0 | -46693 | gtex_brain_putamen_basal |
| rs7302529 | 12 | 77321581 | CSRP2 | ENSG00000175183.5 | 3.63E-9 | 0 | -48741 | gtex_brain_putamen_basal |
| rs35877960 | 12 | 77322751 | CSRP2 | ENSG00000175183.5 | 3.63E-9 | 0 | -49911 | gtex_brain_putamen_basal |
| rs34311394 | 12 | 77323112 | CSRP2 | ENSG00000175183.5 | 3.354E-9 | 0 | -50272 | gtex_brain_putamen_basal |
| rs12823997 | 12 | 77323760 | CSRP2 | ENSG00000175183.5 | 9.909E-8 | 0 | -50920 | gtex_brain_putamen_basal |
| rs1691551 | 12 | 77324116 | CSRP2 | ENSG00000175183.5 | 5.109E-8 | 0 | -51276 | gtex_brain_putamen_basal |
| rs1493428 | 12 | 77326330 | CSRP2 | ENSG00000175183.5 | 1.514E-6 | 0 | -53490 | gtex_brain_putamen_basal |
| rs12813099 | 12 | 77374813 | CSRP2 | ENSG00000175183.5 | 1.829E-6 | 0 | -101973 | gtex_brain_putamen_basal |
| rs11829785 | 12 | 77376271 | CSRP2 | ENSG00000175183.5 | 8.696E-7 | 0 | -103431 | gtex_brain_putamen_basal |
| rs7961235 | 12 | 77377378 | CSRP2 | ENSG00000175183.5 | 8.567E-7 | 0 | -104538 | gtex_brain_putamen_basal |
| rs7976098 | 12 | 77379179 | CSRP2 | ENSG00000175183.5 | 9.817E-7 | 0 | -106339 | gtex_brain_putamen_basal |
| rs7976452 | 12 | 77381447 | CSRP2 | ENSG00000175183.5 | 8.316E-7 | 0 | -108607 | gtex_brain_putamen_basal |
| rs11832777 | 12 | 77383787 | CSRP2 | ENSG00000175183.5 | 8.201E-7 | 0 | -110947 | gtex_brain_putamen_basal |
| rs7963478 | 12 | 77384897 | CSRP2 | ENSG00000175183.5 | 8.161E-7 | 0 | -112057 | gtex_brain_putamen_basal |
| rs10506723 | 12 | 77386509 | CSRP2 | ENSG00000175183.5 | 7.803E-7 | 0 | -113669 | gtex_brain_putamen_basal |
| rs310880 | 12 | 77387885 | CSRP2 | ENSG00000175183.5 | 3.58E-7 | 0 | -115045 | gtex_brain_putamen_basal |
| rs12833647 | 12 | 77391378 | CSRP2 | ENSG00000175183.5 | 6.479E-7 | 0 | -118538 | gtex_brain_putamen_basal |
Section II. Transcriptome annotation
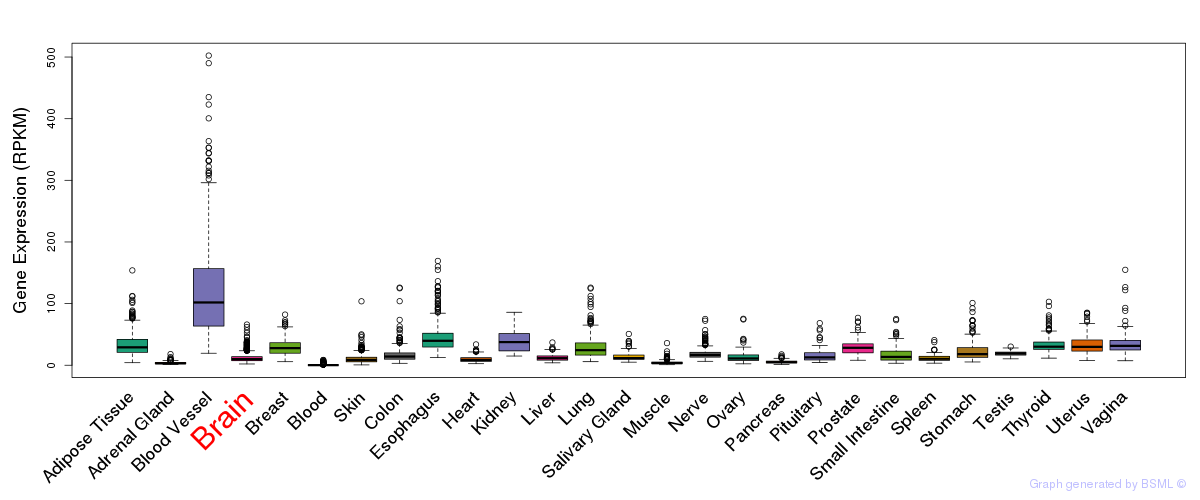
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| RNF34 | 0.93 | 0.93 |
| KIAA0406 | 0.93 | 0.94 |
| C17orf71 | 0.93 | 0.94 |
| KIAA0174 | 0.93 | 0.94 |
| PRPF4 | 0.92 | 0.93 |
| SMU1 | 0.92 | 0.94 |
| EFTUD2 | 0.92 | 0.92 |
| POLR3C | 0.92 | 0.92 |
| BUB3 | 0.92 | 0.95 |
| CSTF2 | 0.92 | 0.91 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| MT-CO2 | -0.83 | -0.86 |
| AF347015.31 | -0.82 | -0.85 |
| AF347015.8 | -0.81 | -0.86 |
| AF347015.33 | -0.81 | -0.84 |
| MT-CYB | -0.80 | -0.84 |
| AF347015.27 | -0.79 | -0.84 |
| AF347015.15 | -0.78 | -0.84 |
| AF347015.2 | -0.77 | -0.83 |
| AF347015.21 | -0.77 | -0.85 |
| FXYD1 | -0.76 | -0.83 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| PID ATF2 PATHWAY | 59 | 43 | All SZGR 2.0 genes in this pathway |
| PARENT MTOR SIGNALING UP | 567 | 375 | All SZGR 2.0 genes in this pathway |
| PICCALUGA ANGIOIMMUNOBLASTIC LYMPHOMA UP | 205 | 140 | All SZGR 2.0 genes in this pathway |
| LIU PROSTATE CANCER DN | 481 | 290 | All SZGR 2.0 genes in this pathway |
| ZHONG RESPONSE TO AZACITIDINE AND TSA UP | 183 | 119 | All SZGR 2.0 genes in this pathway |
| CHARAFE BREAST CANCER LUMINAL VS MESENCHYMAL DN | 460 | 312 | All SZGR 2.0 genes in this pathway |
| LAIHO COLORECTAL CANCER SERRATED UP | 112 | 71 | All SZGR 2.0 genes in this pathway |
| RODRIGUES DCC TARGETS DN | 121 | 84 | All SZGR 2.0 genes in this pathway |
| RHEIN ALL GLUCOCORTICOID THERAPY DN | 362 | 238 | All SZGR 2.0 genes in this pathway |
| PAPASPYRIDONOS UNSTABLE ATEROSCLEROTIC PLAQUE DN | 43 | 29 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC1 TARGETS DN | 260 | 143 | All SZGR 2.0 genes in this pathway |
| TIEN INTESTINE PROBIOTICS 24HR UP | 557 | 331 | All SZGR 2.0 genes in this pathway |
| ELVIDGE HYPOXIA UP | 171 | 112 | All SZGR 2.0 genes in this pathway |
| ELVIDGE HYPOXIA BY DMOG UP | 130 | 85 | All SZGR 2.0 genes in this pathway |
| ELVIDGE HIF1A TARGETS DN | 91 | 58 | All SZGR 2.0 genes in this pathway |
| ELVIDGE HIF1A AND HIF2A TARGETS DN | 104 | 72 | All SZGR 2.0 genes in this pathway |
| DELYS THYROID CANCER DN | 232 | 154 | All SZGR 2.0 genes in this pathway |
| CHIARADONNA NEOPLASTIC TRANSFORMATION KRAS CDC25 UP | 58 | 39 | All SZGR 2.0 genes in this pathway |
| BERENJENO TRANSFORMED BY RHOA DN | 394 | 258 | All SZGR 2.0 genes in this pathway |
| RASHI RESPONSE TO IONIZING RADIATION 2 | 127 | 92 | All SZGR 2.0 genes in this pathway |
| FARMER BREAST CANCER BASAL VS LULMINAL | 330 | 215 | All SZGR 2.0 genes in this pathway |
| WEI MYCN TARGETS WITH E BOX | 795 | 478 | All SZGR 2.0 genes in this pathway |
| ONDER CDH1 TARGETS 1 UP | 140 | 85 | All SZGR 2.0 genes in this pathway |
| LEE LIVER CANCER MYC UP | 54 | 30 | All SZGR 2.0 genes in this pathway |
| LE EGR2 TARGETS UP | 108 | 75 | All SZGR 2.0 genes in this pathway |
| SHIPP DLBCL VS FOLLICULAR LYMPHOMA DN | 45 | 28 | All SZGR 2.0 genes in this pathway |
| PETROVA ENDOTHELIUM LYMPHATIC VS BLOOD UP | 131 | 87 | All SZGR 2.0 genes in this pathway |
| PETROVA PROX1 TARGETS UP | 28 | 16 | All SZGR 2.0 genes in this pathway |
| JAZAERI BREAST CANCER BRCA1 VS BRCA2 DN | 43 | 31 | All SZGR 2.0 genes in this pathway |
| ONGUSAHA TP53 TARGETS | 38 | 23 | All SZGR 2.0 genes in this pathway |
| NIELSEN LIPOSARCOMA DN | 19 | 8 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 48HR DN | 504 | 323 | All SZGR 2.0 genes in this pathway |
| GAJATE RESPONSE TO TRABECTEDIN UP | 67 | 45 | All SZGR 2.0 genes in this pathway |
| BURTON ADIPOGENESIS 3 | 101 | 64 | All SZGR 2.0 genes in this pathway |
| KAAB HEART ATRIUM VS VENTRICLE DN | 261 | 183 | All SZGR 2.0 genes in this pathway |
| KAAB FAILED HEART VENTRICLE DN | 41 | 28 | All SZGR 2.0 genes in this pathway |
| LEIN CHOROID PLEXUS MARKERS | 103 | 61 | All SZGR 2.0 genes in this pathway |
| WANG LSD1 TARGETS UP | 24 | 14 | All SZGR 2.0 genes in this pathway |
| HELLER HDAC TARGETS UP | 317 | 208 | All SZGR 2.0 genes in this pathway |
| HELLER HDAC TARGETS SILENCED BY METHYLATION UP | 461 | 298 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER LUMINAL B DN | 564 | 326 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER BASAL UP | 648 | 398 | All SZGR 2.0 genes in this pathway |
| BREDEMEYER RAG SIGNALING VIA ATM NOT VIA NFKB UP | 49 | 32 | All SZGR 2.0 genes in this pathway |
| VANTVEER BREAST CANCER ESR1 DN | 240 | 153 | All SZGR 2.0 genes in this pathway |
| SWEET LUNG CANCER KRAS UP | 491 | 316 | All SZGR 2.0 genes in this pathway |
| MEISSNER BRAIN HCP WITH H3K4ME3 AND H3K27ME3 | 1069 | 729 | All SZGR 2.0 genes in this pathway |
| YAGI AML WITH T 8 21 TRANSLOCATION | 368 | 247 | All SZGR 2.0 genes in this pathway |
| CAIRO HEPATOBLASTOMA CLASSES DN | 210 | 141 | All SZGR 2.0 genes in this pathway |
| CAIRO LIVER DEVELOPMENT UP | 166 | 105 | All SZGR 2.0 genes in this pathway |
| WONG EMBRYONIC STEM CELL CORE | 335 | 193 | All SZGR 2.0 genes in this pathway |
| YAO TEMPORAL RESPONSE TO PROGESTERONE CLUSTER 13 | 172 | 107 | All SZGR 2.0 genes in this pathway |
| FEVR CTNNB1 TARGETS DN | 553 | 343 | All SZGR 2.0 genes in this pathway |
| GOBERT OLIGODENDROCYTE DIFFERENTIATION UP | 570 | 339 | All SZGR 2.0 genes in this pathway |
| KRIEG HYPOXIA NOT VIA KDM3A | 770 | 480 | All SZGR 2.0 genes in this pathway |
| LIM MAMMARY STEM CELL UP | 489 | 314 | All SZGR 2.0 genes in this pathway |