Gene Page: CYP2E1
Summary ?
| GeneID | 1571 |
| Symbol | CYP2E1 |
| Synonyms | CPE1|CYP2E|P450-J|P450C2E |
| Description | cytochrome P450 family 2 subfamily E member 1 |
| Reference | MIM:124040|HGNC:HGNC:2631|Ensembl:ENSG00000130649|HPRD:11813|Vega:OTTHUMG00000019322 |
| Gene type | protein-coding |
| Map location | 10q26.3 |
| Pascal p-value | 0.079 |
| Sherlock p-value | 0.717 |
| Fetal beta | -0.661 |
| eGene | Cortex Putamen basal ganglia Myers' cis & trans Meta |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs10857731 | chr10 | 135328359 | CYP2E1 | 1571 | 0.14 | cis | ||
| rs633915 | chr1 | 182682157 | CYP2E1 | 1571 | 0.15 | trans | ||
| rs621071 | chr1 | 182682810 | CYP2E1 | 1571 | 0 | trans | ||
| rs9419072 | 10 | 135310445 | CYP2E1 | ENSG00000130649.5 | 1.02E-6 | 0 | -23465 | gtex_brain_putamen_basal |
| rs1854458 | 10 | 135313815 | CYP2E1 | ENSG00000130649.5 | 1.02E-6 | 0 | -20095 | gtex_brain_putamen_basal |
| rs9418982 | 10 | 135320622 | CYP2E1 | ENSG00000130649.5 | 1.02E-6 | 0 | -13288 | gtex_brain_putamen_basal |
| rs9418984 | 10 | 135322356 | CYP2E1 | ENSG00000130649.5 | 2.506E-6 | 0 | -11554 | gtex_brain_putamen_basal |
| rs7083073 | 10 | 135322956 | CYP2E1 | ENSG00000130649.5 | 5.052E-9 | 0 | -10954 | gtex_brain_putamen_basal |
| rs7092634 | 10 | 135322977 | CYP2E1 | ENSG00000130649.5 | 5.052E-9 | 0 | -10933 | gtex_brain_putamen_basal |
| rs6537609 | 10 | 135323084 | CYP2E1 | ENSG00000130649.5 | 5.052E-9 | 0 | -10826 | gtex_brain_putamen_basal |
| rs7911139 | 10 | 135323525 | CYP2E1 | ENSG00000130649.5 | 4.576E-9 | 0 | -10385 | gtex_brain_putamen_basal |
| rs7077457 | 10 | 135324370 | CYP2E1 | ENSG00000130649.5 | 5.052E-9 | 0 | -9540 | gtex_brain_putamen_basal |
| rs9418987 | 10 | 135324409 | CYP2E1 | ENSG00000130649.5 | 2.483E-6 | 0 | -9501 | gtex_brain_putamen_basal |
| rs7096203 | 10 | 135324505 | CYP2E1 | ENSG00000130649.5 | 5.052E-9 | 0 | -9405 | gtex_brain_putamen_basal |
| rs7096321 | 10 | 135324536 | CYP2E1 | ENSG00000130649.5 | 5.052E-9 | 0 | -9374 | gtex_brain_putamen_basal |
| rs10857730 | 10 | 135326822 | CYP2E1 | ENSG00000130649.5 | 4.311E-9 | 0 | -7088 | gtex_brain_putamen_basal |
| rs9419079 | 10 | 135326985 | CYP2E1 | ENSG00000130649.5 | 1.016E-7 | 0 | -6925 | gtex_brain_putamen_basal |
| rs10776685 | 10 | 135327317 | CYP2E1 | ENSG00000130649.5 | 5.052E-9 | 0 | -6593 | gtex_brain_putamen_basal |
| rs1581934 | 10 | 135327410 | CYP2E1 | ENSG00000130649.5 | 5.052E-9 | 0 | -6500 | gtex_brain_putamen_basal |
| rs7095379 | 10 | 135328532 | CYP2E1 | ENSG00000130649.5 | 5.052E-9 | 0 | -5378 | gtex_brain_putamen_basal |
| rs7095996 | 10 | 135328948 | CYP2E1 | ENSG00000130649.5 | 5.052E-9 | 0 | -4962 | gtex_brain_putamen_basal |
| rs11815623 | 10 | 135329133 | CYP2E1 | ENSG00000130649.5 | 5.027E-9 | 0 | -4777 | gtex_brain_putamen_basal |
| rs2026047 | 10 | 135329616 | CYP2E1 | ENSG00000130649.5 | 2.149E-6 | 0 | -4294 | gtex_brain_putamen_basal |
| rs751945 | 10 | 135329939 | CYP2E1 | ENSG00000130649.5 | 1.974E-9 | 0 | -3971 | gtex_brain_putamen_basal |
| rs751946 | 10 | 135330104 | CYP2E1 | ENSG00000130649.5 | 2.701E-12 | 0 | -3806 | gtex_brain_putamen_basal |
| rs9418990 | 10 | 135337966 | CYP2E1 | ENSG00000130649.5 | 6.895E-7 | 0 | 4056 | gtex_brain_putamen_basal |
| rs2070673 | 10 | 135340567 | CYP2E1 | ENSG00000130649.5 | 5.563E-7 | 0 | 6657 | gtex_brain_putamen_basal |
| rs1536828 | 10 | 135342315 | CYP2E1 | ENSG00000130649.5 | 2.315E-6 | 0 | 8405 | gtex_brain_putamen_basal |
| rs8192770 | 10 | 135342644 | CYP2E1 | ENSG00000130649.5 | 3.089E-6 | 0 | 8734 | gtex_brain_putamen_basal |
| rs8192776 | 10 | 135348187 | CYP2E1 | ENSG00000130649.5 | 3.366E-6 | 0 | 14277 | gtex_brain_putamen_basal |
| rs2480257 | 10 | 135352509 | CYP2E1 | ENSG00000130649.5 | 2.871E-6 | 0 | 18599 | gtex_brain_putamen_basal |
| rs4363513 | 10 | 135355534 | CYP2E1 | ENSG00000130649.5 | 1.779E-7 | 0 | 21624 | gtex_brain_putamen_basal |
| rs4240522 | 10 | 135355557 | CYP2E1 | ENSG00000130649.5 | 1.7E-7 | 0 | 21647 | gtex_brain_putamen_basal |
| rs1536826 | 10 | 135357239 | CYP2E1 | ENSG00000130649.5 | 3.352E-7 | 0 | 23329 | gtex_brain_putamen_basal |
| rs3020513 | 10 | 135364915 | CYP2E1 | ENSG00000130649.5 | 1.076E-6 | 0 | 31005 | gtex_brain_putamen_basal |
| rs2987785 | 10 | 135365003 | CYP2E1 | ENSG00000130649.5 | 6.522E-7 | 0 | 31093 | gtex_brain_putamen_basal |
Section II. Transcriptome annotation
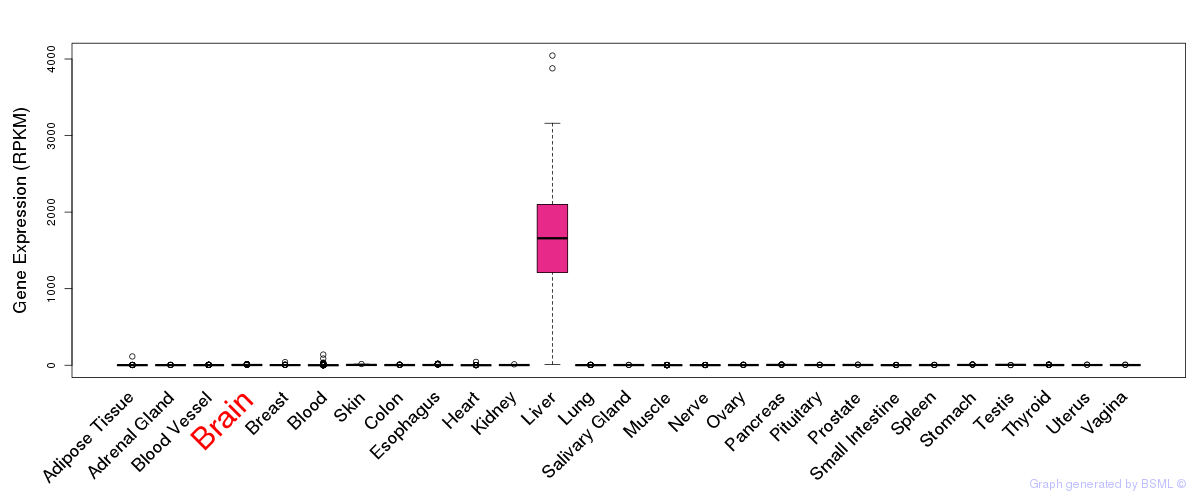
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| TNFSF13 | 0.87 | 0.81 |
| GPR143 | 0.86 | 0.82 |
| PRDX6 | 0.84 | 0.76 |
| GJB6 | 0.84 | 0.81 |
| TAPBPL | 0.83 | 0.78 |
| SNTA1 | 0.82 | 0.81 |
| TMBIM1 | 0.81 | 0.81 |
| NDRG2 | 0.81 | 0.84 |
| AGXT2L1 | 0.81 | 0.82 |
| EPHX1 | 0.80 | 0.80 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| BLMH | -0.57 | -0.69 |
| RTF1 | -0.57 | -0.68 |
| NOVA2 | -0.56 | -0.70 |
| RNF40 | -0.56 | -0.69 |
| HEATR5B | -0.56 | -0.67 |
| NKIRAS2 | -0.56 | -0.71 |
| PJA1 | -0.56 | -0.69 |
| NOL4 | -0.55 | -0.70 |
| BASP1 | -0.55 | -0.68 |
| TOPBP1 | -0.55 | -0.66 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG ARACHIDONIC ACID METABOLISM | 58 | 36 | All SZGR 2.0 genes in this pathway |
| KEGG LINOLEIC ACID METABOLISM | 29 | 16 | All SZGR 2.0 genes in this pathway |
| KEGG METABOLISM OF XENOBIOTICS BY CYTOCHROME P450 | 70 | 44 | All SZGR 2.0 genes in this pathway |
| KEGG DRUG METABOLISM CYTOCHROME P450 | 72 | 47 | All SZGR 2.0 genes in this pathway |
| BIOCARTA NUCLEARRS PATHWAY | 15 | 12 | All SZGR 2.0 genes in this pathway |
| REACTOME BIOLOGICAL OXIDATIONS | 139 | 91 | All SZGR 2.0 genes in this pathway |
| REACTOME XENOBIOTICS | 16 | 11 | All SZGR 2.0 genes in this pathway |
| REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | 51 | 34 | All SZGR 2.0 genes in this pathway |
| REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | 70 | 50 | All SZGR 2.0 genes in this pathway |
| GAZDA DIAMOND BLACKFAN ANEMIA ERYTHROID DN | 493 | 298 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN MLL SIGNATURE 1 DN | 242 | 165 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN MLL SIGNATURE 2 DN | 281 | 186 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA UP | 1821 | 933 | All SZGR 2.0 genes in this pathway |
| LASTOWSKA NEUROBLASTOMA COPY NUMBER DN | 800 | 473 | All SZGR 2.0 genes in this pathway |
| RICKMAN HEAD AND NECK CANCER E | 89 | 44 | All SZGR 2.0 genes in this pathway |
| LEE LIVER CANCER CIPROFIBRATE DN | 66 | 43 | All SZGR 2.0 genes in this pathway |
| LEE LIVER CANCER MYC E2F1 DN | 64 | 38 | All SZGR 2.0 genes in this pathway |
| LEE LIVER CANCER DENA DN | 74 | 45 | All SZGR 2.0 genes in this pathway |
| LEE LIVER CANCER E2F1 DN | 64 | 42 | All SZGR 2.0 genes in this pathway |
| YAO HOXA10 TARGETS VIA PROGESTERONE UP | 79 | 58 | All SZGR 2.0 genes in this pathway |
| HSIAO LIVER SPECIFIC GENES | 244 | 154 | All SZGR 2.0 genes in this pathway |
| NADLER OBESITY DN | 48 | 34 | All SZGR 2.0 genes in this pathway |
| NGUYEN NOTCH1 TARGETS DN | 86 | 67 | All SZGR 2.0 genes in this pathway |
| VERHAAK AML WITH NPM1 MUTATED DN | 246 | 180 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS STEM CELL | 254 | 164 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 18HR UP | 178 | 111 | All SZGR 2.0 genes in this pathway |
| JACKSON DNMT1 TARGETS UP | 77 | 57 | All SZGR 2.0 genes in this pathway |
| TUOMISTO TUMOR SUPPRESSION BY COL13A1 DN | 17 | 11 | All SZGR 2.0 genes in this pathway |
| LEE LIVER CANCER | 49 | 29 | All SZGR 2.0 genes in this pathway |
| IWANAGA CARCINOGENESIS BY KRAS DN | 120 | 81 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER CANCER DN | 540 | 340 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER TUMOR VS NORMAL ADJACENT TISSUE DN | 274 | 165 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER CANCER WITH H3K27ME3 UP | 295 | 149 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER CANCER WITH H3K9ME3 UP | 141 | 75 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER BASAL DN | 701 | 446 | All SZGR 2.0 genes in this pathway |
| BOCHKIS FOXA2 TARGETS | 425 | 261 | All SZGR 2.0 genes in this pathway |
| CADWELL ATG16L1 TARGETS UP | 93 | 56 | All SZGR 2.0 genes in this pathway |
| MOOTHA MITOCHONDRIA | 447 | 277 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS UP | 1305 | 895 | All SZGR 2.0 genes in this pathway |
| WUNDER INFLAMMATORY RESPONSE AND CHOLESTEROL DN | 12 | 7 | All SZGR 2.0 genes in this pathway |
| SWEET LUNG CANCER KRAS DN | 435 | 289 | All SZGR 2.0 genes in this pathway |
| SHEDDEN LUNG CANCER GOOD SURVIVAL A12 | 317 | 177 | All SZGR 2.0 genes in this pathway |
| CHEN METABOLIC SYNDROM NETWORK | 1210 | 725 | All SZGR 2.0 genes in this pathway |
| CHIANG LIVER CANCER SUBCLASS CTNNB1 UP | 176 | 110 | All SZGR 2.0 genes in this pathway |
| WOO LIVER CANCER RECURRENCE DN | 80 | 54 | All SZGR 2.0 genes in this pathway |
| YAGI AML WITH INV 16 TRANSLOCATION | 422 | 277 | All SZGR 2.0 genes in this pathway |
| YAGI AML WITH T 9 11 TRANSLOCATION | 130 | 87 | All SZGR 2.0 genes in this pathway |
| CAIRO HEPATOBLASTOMA CLASSES DN | 210 | 141 | All SZGR 2.0 genes in this pathway |
| CAIRO HEPATOBLASTOMA POOR SURVIVAL | 16 | 9 | All SZGR 2.0 genes in this pathway |
| YAO TEMPORAL RESPONSE TO PROGESTERONE CLUSTER 5 | 30 | 22 | All SZGR 2.0 genes in this pathway |