Gene Page: CYP4B1
Summary ?
| GeneID | 1580 |
| Symbol | CYP4B1 |
| Synonyms | CYPIVB1|P-450HP |
| Description | cytochrome P450 family 4 subfamily B member 1 |
| Reference | MIM:124075|HGNC:HGNC:2644|Ensembl:ENSG00000142973|HPRD:00491|Vega:OTTHUMG00000007984 |
| Gene type | protein-coding |
| Map location | 1p33 |
| Pascal p-value | 0.175 |
| Fetal beta | -0.118 |
| eGene | Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs17131472 | chr1 | 91962966 | CYP4B1 | 1580 | 0.06 | trans | ||
| rs3762382 | chr1 | 198128858 | CYP4B1 | 1580 | 0.12 | trans | ||
| rs6546473 | chr2 | 69260356 | CYP4B1 | 1580 | 0.07 | trans | ||
| rs1566993 | chr2 | 117105059 | CYP4B1 | 1580 | 0.13 | trans | ||
| rs10050805 | chr5 | 30805352 | CYP4B1 | 1580 | 0.17 | trans | ||
| rs9381511 | chr6 | 12931447 | CYP4B1 | 1580 | 0.07 | trans | ||
| rs6455226 | chr6 | 67930295 | CYP4B1 | 1580 | 0 | trans | ||
| rs7081666 | chr10 | 47673800 | CYP4B1 | 1580 | 0.1 | trans | ||
| rs10733768 | chr10 | 52667246 | CYP4B1 | 1580 | 0.02 | trans | ||
| rs1159612 | chr10 | 85329057 | CYP4B1 | 1580 | 0.01 | trans | ||
| rs10879027 | chr12 | 70237156 | CYP4B1 | 1580 | 0.02 | trans | ||
| rs16958302 | chr16 | 57647432 | CYP4B1 | 1580 | 0.06 | trans | ||
| rs6506551 | chr18 | 8167049 | CYP4B1 | 1580 | 0.18 | trans | ||
| rs4805564 | chr19 | 30911055 | CYP4B1 | 1580 | 0.04 | trans | ||
| rs6095741 | chr20 | 48666589 | CYP4B1 | 1580 | 2.007E-8 | trans | ||
| rs5915713 | chrX | 4077883 | CYP4B1 | 1580 | 0.02 | trans | ||
| rs5991662 | chrX | 43335796 | CYP4B1 | 1580 | 0 | trans | ||
| rs1545676 | chrX | 93530472 | CYP4B1 | 1580 | 1.266E-4 | trans | ||
| rs242163 | chrX | 132311662 | CYP4B1 | 1580 | 0.06 | trans | ||
| rs17340148 | chrX | 138648246 | CYP4B1 | 1580 | 0 | trans | ||
| rs10521801 | chrX | 138732866 | CYP4B1 | 1580 | 0.06 | trans |
Section II. Transcriptome annotation
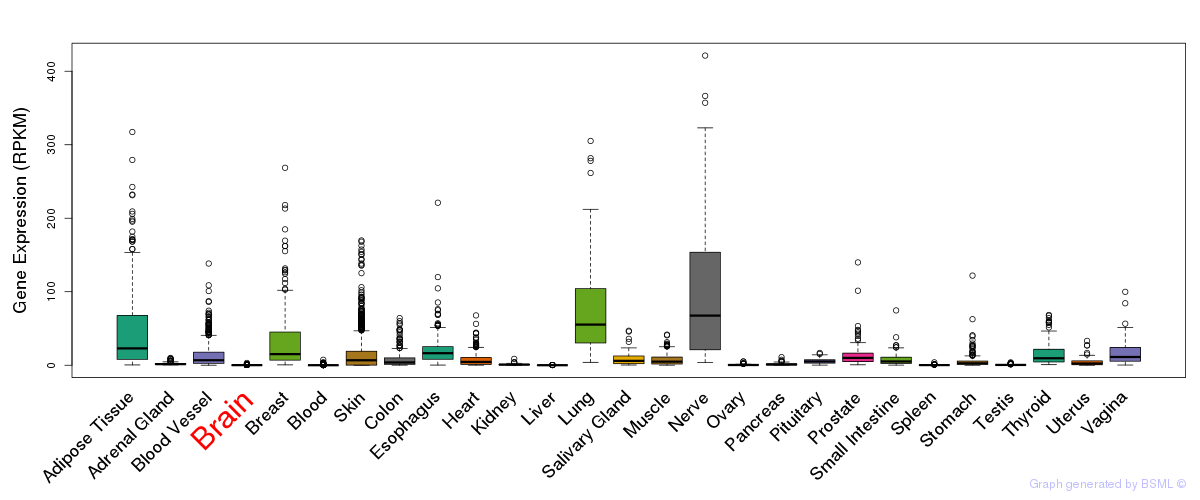
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| CSRP2 | 0.83 | 0.55 |
| AP000926.2 | 0.82 | 0.71 |
| SLA | 0.80 | 0.78 |
| C8orf79 | 0.79 | 0.73 |
| PTPRO | 0.79 | 0.75 |
| POU3F2 | 0.77 | 0.68 |
| DOPEY2 | 0.77 | 0.70 |
| KRT31 | 0.77 | 0.72 |
| VSTM2L | 0.76 | 0.75 |
| SEMA3C | 0.76 | 0.74 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| HEPN1 | -0.54 | -0.68 |
| HLA-F | -0.53 | -0.62 |
| RENBP | -0.53 | -0.67 |
| AIFM3 | -0.53 | -0.66 |
| CLU | -0.53 | -0.62 |
| TSC22D4 | -0.52 | -0.67 |
| AP003117.1 | -0.52 | -0.58 |
| FAH | -0.52 | -0.66 |
| HSD17B14 | -0.52 | -0.67 |
| BDH2 | -0.52 | -0.69 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| REACTOME BIOLOGICAL OXIDATIONS | 139 | 91 | All SZGR 2.0 genes in this pathway |
| REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | 51 | 34 | All SZGR 2.0 genes in this pathway |
| REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | 70 | 50 | All SZGR 2.0 genes in this pathway |
| LIU PROSTATE CANCER DN | 481 | 290 | All SZGR 2.0 genes in this pathway |
| SENGUPTA NASOPHARYNGEAL CARCINOMA DN | 349 | 157 | All SZGR 2.0 genes in this pathway |
| THUM SYSTOLIC HEART FAILURE DN | 244 | 147 | All SZGR 2.0 genes in this pathway |
| CHARAFE BREAST CANCER LUMINAL VS MESENCHYMAL UP | 450 | 256 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA UP | 1821 | 933 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE EPIDERMIS DN | 508 | 354 | All SZGR 2.0 genes in this pathway |
| LANDIS ERBB2 BREAST TUMORS 324 DN | 149 | 93 | All SZGR 2.0 genes in this pathway |
| SAENZ DETOX PATHWAY AND CARCINOGENESIS DN | 13 | 11 | All SZGR 2.0 genes in this pathway |
| LIN SILENCED BY TUMOR MICROENVIRONMENT | 108 | 73 | All SZGR 2.0 genes in this pathway |
| SUZUKI RESPONSE TO TSA AND DECITABINE 1A | 23 | 16 | All SZGR 2.0 genes in this pathway |
| CUI TCF21 TARGETS 2 DN | 830 | 547 | All SZGR 2.0 genes in this pathway |
| KEEN RESPONSE TO ROSIGLITAZONE UP | 38 | 23 | All SZGR 2.0 genes in this pathway |
| CHEN LVAD SUPPORT OF FAILING HEART UP | 103 | 69 | All SZGR 2.0 genes in this pathway |
| HENDRICKS SMARCA4 TARGETS DN | 53 | 34 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 TARGETS UP | 673 | 430 | All SZGR 2.0 genes in this pathway |
| MARTINEZ TP53 TARGETS UP | 602 | 364 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 AND TP53 TARGETS UP | 601 | 369 | All SZGR 2.0 genes in this pathway |
| SARRIO EPITHELIAL MESENCHYMAL TRANSITION DN | 154 | 101 | All SZGR 2.0 genes in this pathway |
| IWANAGA CARCINOGENESIS BY KRAS DN | 120 | 81 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER RELAPSE IN BRAIN DN | 85 | 58 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER LUMINAL A UP | 84 | 52 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER BASAL DN | 701 | 446 | All SZGR 2.0 genes in this pathway |
| BOQUEST STEM CELL CULTURED VS FRESH UP | 425 | 298 | All SZGR 2.0 genes in this pathway |
| SWEET LUNG CANCER KRAS DN | 435 | 289 | All SZGR 2.0 genes in this pathway |
| POOLA INVASIVE BREAST CANCER DN | 134 | 83 | All SZGR 2.0 genes in this pathway |
| MARTENS TRETINOIN RESPONSE UP | 857 | 456 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE VIA TP53 GROUP A | 898 | 516 | All SZGR 2.0 genes in this pathway |
| PEDERSEN METASTASIS BY ERBB2 ISOFORM 7 | 403 | 240 | All SZGR 2.0 genes in this pathway |