Gene Page: CYP19A1
Summary ?
| GeneID | 1588 |
| Symbol | CYP19A1 |
| Synonyms | ARO|ARO1|CPV1|CYAR|CYP19|CYPXIX|P-450AROM |
| Description | cytochrome P450 family 19 subfamily A member 1 |
| Reference | MIM:107910|HGNC:HGNC:2594|Ensembl:ENSG00000137869|HPRD:00488|Vega:OTTHUMG00000131747 |
| Gene type | protein-coding |
| Map location | 15q21.1 |
| Pascal p-value | 8.712E-4 |
| Fetal beta | -0.268 |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| CV:PheWAS | Phenome-wide association studies (PheWAS) | 157 SNPs associated with schizophrenia | 1 |
Section I. Genetics and epigenetics annotation
 CV:PheWAS
CV:PheWAS
| SNP ID | Chromosome | Position | Allele | P | Function | Gene | Up/Down Distance |
|---|---|---|---|---|---|---|---|
| rs2305707 | 15 | 51569410 | null | 1.555 | CYP19A1 | null |
Section II. Transcriptome annotation
General gene expression (GTEx)
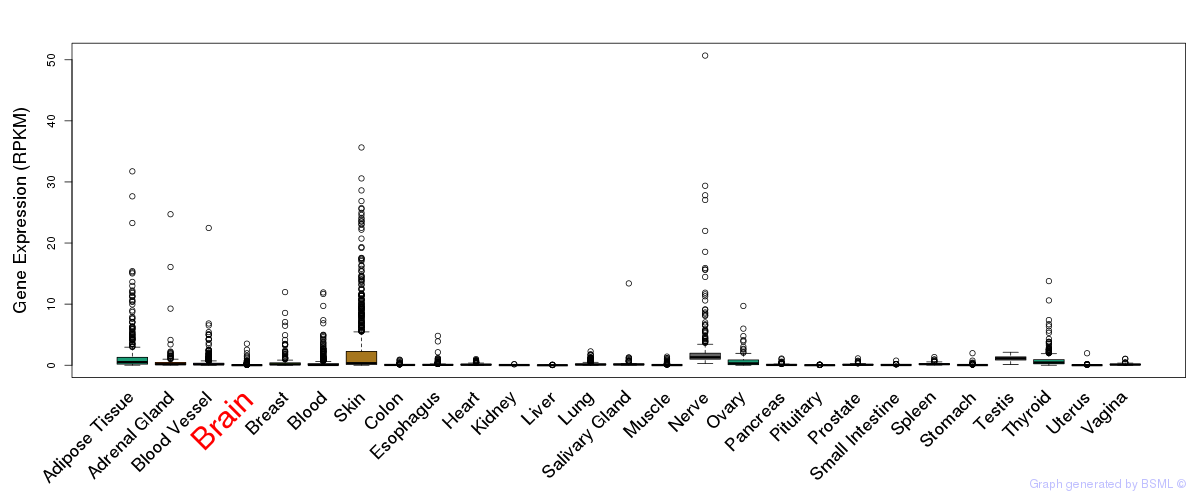
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| EML1 | 0.96 | 0.87 |
| AFF3 | 0.95 | 0.91 |
| GPR12 | 0.94 | 0.87 |
| FAT4 | 0.94 | 0.91 |
| FAM117B | 0.93 | 0.90 |
| FAM59A | 0.93 | 0.87 |
| SSBP2 | 0.93 | 0.89 |
| SEZ6 | 0.93 | 0.90 |
| BACH2 | 0.93 | 0.89 |
| DACT1 | 0.92 | 0.94 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| SERPINB6 | -0.65 | -0.75 |
| HEPN1 | -0.61 | -0.78 |
| LHPP | -0.61 | -0.61 |
| C5orf53 | -0.61 | -0.75 |
| AIFM3 | -0.60 | -0.80 |
| HLA-F | -0.60 | -0.79 |
| TNFSF12 | -0.60 | -0.59 |
| FBXO2 | -0.60 | -0.65 |
| HSD17B14 | -0.60 | -0.80 |
| TSC22D4 | -0.59 | -0.81 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG STEROID HORMONE BIOSYNTHESIS | 55 | 30 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | 35 | 23 | All SZGR 2.0 genes in this pathway |
| REACTOME BIOLOGICAL OXIDATIONS | 139 | 91 | All SZGR 2.0 genes in this pathway |
| REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | 51 | 34 | All SZGR 2.0 genes in this pathway |
| REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | 70 | 50 | All SZGR 2.0 genes in this pathway |
| REACTOME ENDOGENOUS STEROLS | 15 | 10 | All SZGR 2.0 genes in this pathway |
| REACTOME STEROID HORMONES | 29 | 21 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF LIPIDS AND LIPOPROTEINS | 478 | 302 | All SZGR 2.0 genes in this pathway |
| PARENT MTOR SIGNALING UP | 567 | 375 | All SZGR 2.0 genes in this pathway |
| ODONNELL TFRC TARGETS UP | 456 | 228 | All SZGR 2.0 genes in this pathway |
| GOZGIT ESR1 TARGETS DN | 781 | 465 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE KERATINOCYTE UP | 530 | 342 | All SZGR 2.0 genes in this pathway |
| MOHANKUMAR TLX1 TARGETS DN | 193 | 112 | All SZGR 2.0 genes in this pathway |
| ASTON MAJOR DEPRESSIVE DISORDER DN | 160 | 110 | All SZGR 2.0 genes in this pathway |
| MAHAJAN RESPONSE TO IL1A DN | 76 | 57 | All SZGR 2.0 genes in this pathway |
| GAJATE RESPONSE TO TRABECTEDIN UP | 67 | 45 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 18HR UP | 178 | 111 | All SZGR 2.0 genes in this pathway |
| LEE CALORIE RESTRICTION NEOCORTEX DN | 88 | 58 | All SZGR 2.0 genes in this pathway |
| BANDRES RESPONSE TO CARMUSTIN MGMT 48HR DN | 161 | 105 | All SZGR 2.0 genes in this pathway |
| KRASNOSELSKAYA ILF3 TARGETS DN | 46 | 38 | All SZGR 2.0 genes in this pathway |
| SU PLACENTA | 30 | 18 | All SZGR 2.0 genes in this pathway |
| KYNG DNA DAMAGE BY 4NQO OR GAMMA RADIATION | 15 | 13 | All SZGR 2.0 genes in this pathway |
| KYNG DNA DAMAGE UP | 226 | 164 | All SZGR 2.0 genes in this pathway |
| KONDO EZH2 TARGETS | 245 | 148 | All SZGR 2.0 genes in this pathway |
| LEE METASTASIS AND ALTERNATIVE SPLICING DN | 45 | 31 | All SZGR 2.0 genes in this pathway |
| MATZUK STEROIDOGENESIS | 7 | 6 | All SZGR 2.0 genes in this pathway |
| MATZUK MALE REPRODUCTION SERTOLI | 28 | 23 | All SZGR 2.0 genes in this pathway |
| MATZUK SPERMATOGONIA | 24 | 17 | All SZGR 2.0 genes in this pathway |
| ALONSO METASTASIS DN | 26 | 17 | All SZGR 2.0 genes in this pathway |
| SASSON RESPONSE TO GONADOTROPHINS UP | 91 | 59 | All SZGR 2.0 genes in this pathway |
| SASSON RESPONSE TO FORSKOLIN UP | 90 | 58 | All SZGR 2.0 genes in this pathway |
| SUMI HNF4A TARGETS | 34 | 14 | All SZGR 2.0 genes in this pathway |