Gene Page: ADSS
Summary ?
| GeneID | 159 |
| Symbol | ADSS |
| Synonyms | ADEH|ADSS 2 |
| Description | adenylosuccinate synthase |
| Reference | MIM:103060|HGNC:HGNC:292|Ensembl:ENSG00000035687|HPRD:00050|Vega:OTTHUMG00000040102 |
| Gene type | protein-coding |
| Map location | 1q44 |
| Fetal beta | -0.08 |
| DMG | 2 (# studies) |
| eGene | Meta |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 2 |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 2 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenics,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg01012879 | 1 | 244613363 | ADSS | 4.14E-5 | -0.582 | 0.021 | DMG:Wockner_2014 |
| cg19375676 | 1 | 244615173 | ADSS | 4.73E-9 | -0.009 | 2.73E-6 | DMG:Jaffe_2016 |
Section II. Transcriptome annotation
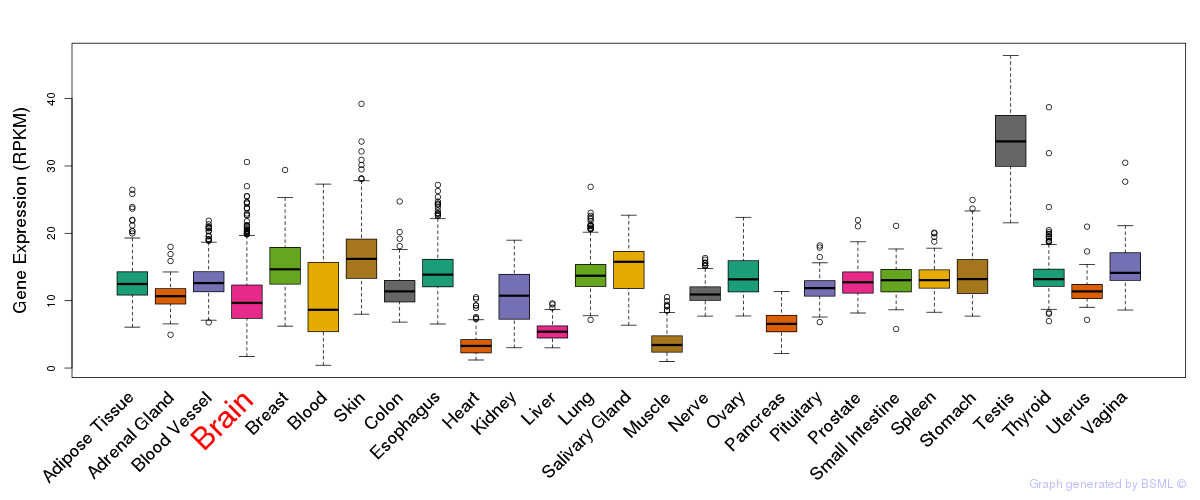
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| ZDHHC5 | 0.92 | 0.93 |
| DGCR2 | 0.92 | 0.93 |
| ALKBH5 | 0.90 | 0.90 |
| AP2A1 | 0.90 | 0.89 |
| NOL6 | 0.88 | 0.90 |
| ARHGEF11 | 0.88 | 0.89 |
| RPTOR | 0.88 | 0.91 |
| TRAPPC9 | 0.88 | 0.88 |
| PVR | 0.88 | 0.89 |
| C2orf18 | 0.88 | 0.88 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.21 | -0.73 | -0.73 |
| AF347015.31 | -0.67 | -0.65 |
| MT-CO2 | -0.65 | -0.65 |
| C1orf54 | -0.65 | -0.69 |
| GNG11 | -0.64 | -0.63 |
| AF347015.8 | -0.63 | -0.63 |
| SYCP3 | -0.62 | -0.69 |
| AL050337.1 | -0.62 | -0.68 |
| AP002478.3 | -0.61 | -0.61 |
| HIGD1B | -0.61 | -0.61 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0000166 | nucleotide binding | IEA | - | |
| GO:0000287 | magnesium ion binding | IEA | - | |
| GO:0004019 | adenylosuccinate synthase activity | IDA | 2004783 | |
| GO:0004019 | adenylosuccinate synthase activity | IEA | - | |
| GO:0004019 | adenylosuccinate synthase activity | NAS | 1592113 | |
| GO:0005525 | GTP binding | IEA | - | |
| GO:0005525 | GTP binding | NAS | 1592113 |15786719 | |
| GO:0016874 | ligase activity | IEA | - | |
| GO:0042301 | phosphate binding | NAS | 1592113 |15786719 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0002376 | immune system process | NAS | 15786719 | |
| GO:0006167 | AMP biosynthetic process | IDA | 2004783 | |
| GO:0006167 | AMP biosynthetic process | NAS | 1592113 |15786719 | |
| GO:0006164 | purine nucleotide biosynthetic process | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005575 | cellular_component | ND | - | |
| GO:0005737 | cytoplasm | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG PURINE METABOLISM | 159 | 96 | All SZGR 2.0 genes in this pathway |
| KEGG ALANINE ASPARTATE AND GLUTAMATE METABOLISM | 32 | 26 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF NUCLEOTIDES | 72 | 45 | All SZGR 2.0 genes in this pathway |
| REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | 11 | 8 | All SZGR 2.0 genes in this pathway |
| REACTOME PURINE METABOLISM | 33 | 24 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP | 1382 | 904 | All SZGR 2.0 genes in this pathway |
| OSMAN BLADDER CANCER UP | 404 | 246 | All SZGR 2.0 genes in this pathway |
| KINSEY TARGETS OF EWSR1 FLII FUSION UP | 1278 | 748 | All SZGR 2.0 genes in this pathway |
| KIM WT1 TARGETS DN | 459 | 276 | All SZGR 2.0 genes in this pathway |
| SCHLOSSER MYC TARGETS REPRESSED BY SERUM | 159 | 93 | All SZGR 2.0 genes in this pathway |
| GRUETZMANN PANCREATIC CANCER UP | 358 | 245 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA1 PCC NETWORK | 1652 | 1023 | All SZGR 2.0 genes in this pathway |
| NUYTTEN NIPP1 TARGETS UP | 769 | 437 | All SZGR 2.0 genes in this pathway |
| NUYTTEN EZH2 TARGETS UP | 1037 | 673 | All SZGR 2.0 genes in this pathway |
| GARCIA TARGETS OF FLI1 AND DAX1 DN | 176 | 104 | All SZGR 2.0 genes in this pathway |
| SHEN SMARCA2 TARGETS UP | 424 | 268 | All SZGR 2.0 genes in this pathway |
| TCGA GLIOBLASTOMA COPY NUMBER UP | 75 | 36 | All SZGR 2.0 genes in this pathway |
| LIN APC TARGETS | 77 | 55 | All SZGR 2.0 genes in this pathway |
| BYSTRYKH HEMATOPOIESIS STEM CELL QTL TRANS | 882 | 572 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS LATE PROGENITOR | 544 | 307 | All SZGR 2.0 genes in this pathway |
| MARIADASON REGULATED BY HISTONE ACETYLATION UP | 83 | 49 | All SZGR 2.0 genes in this pathway |
| WELCSH BRCA1 TARGETS UP | 198 | 132 | All SZGR 2.0 genes in this pathway |
| GAVIN FOXP3 TARGETS CLUSTER T4 | 94 | 69 | All SZGR 2.0 genes in this pathway |
| ZHENG BOUND BY FOXP3 | 491 | 310 | All SZGR 2.0 genes in this pathway |
| RIZKI TUMOR INVASIVENESS 3D UP | 210 | 124 | All SZGR 2.0 genes in this pathway |
| MOOTHA PGC | 420 | 269 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS DN | 1972 | 1213 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-19 | 392 | 399 | 1A,m8 | hsa-miR-19a | UGUGCAAAUCUAUGCAAAACUGA |
| hsa-miR-19b | UGUGCAAAUCCAUGCAAAACUGA | ||||
| miR-214 | 533 | 539 | 1A | hsa-miR-214brain | ACAGCAGGCACAGACAGGCAG |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.