Gene Page: CYP24A1
Summary ?
| GeneID | 1591 |
| Symbol | CYP24A1 |
| Synonyms | CP24|CYP24|HCAI|P450-CC24 |
| Description | cytochrome P450 family 24 subfamily A member 1 |
| Reference | MIM:126065|HGNC:HGNC:2602|Ensembl:ENSG00000019186|HPRD:00522|Vega:OTTHUMG00000032773 |
| Gene type | protein-coding |
| Map location | 20q13 |
| Pascal p-value | 0.282 |
| Fetal beta | -1.203 |
| eGene | Anterior cingulate cortex BA24 Cerebellum Cortex Frontal Cortex BA9 Nucleus accumbens basal ganglia |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs6022999 | 20 | 52788013 | CYP24A1 | ENSG00000019186.5 | 5.82957E-7 | 0 | 2499 | gtex_brain_ba24 |
| rs2259735 | 20 | 52788314 | CYP24A1 | ENSG00000019186.5 | 6.46602E-8 | 0 | 2198 | gtex_brain_ba24 |
| rs2248137 | 20 | 52789743 | CYP24A1 | ENSG00000019186.5 | 6.1939E-9 | 0 | 769 | gtex_brain_ba24 |
| rs2585427 | 20 | 52790976 | CYP24A1 | ENSG00000019186.5 | 4.07207E-7 | 0 | -464 | gtex_brain_ba24 |
| rs2248359 | 20 | 52791518 | CYP24A1 | ENSG00000019186.5 | 7.80643E-9 | 0 | -1006 | gtex_brain_ba24 |
Section II. Transcriptome annotation
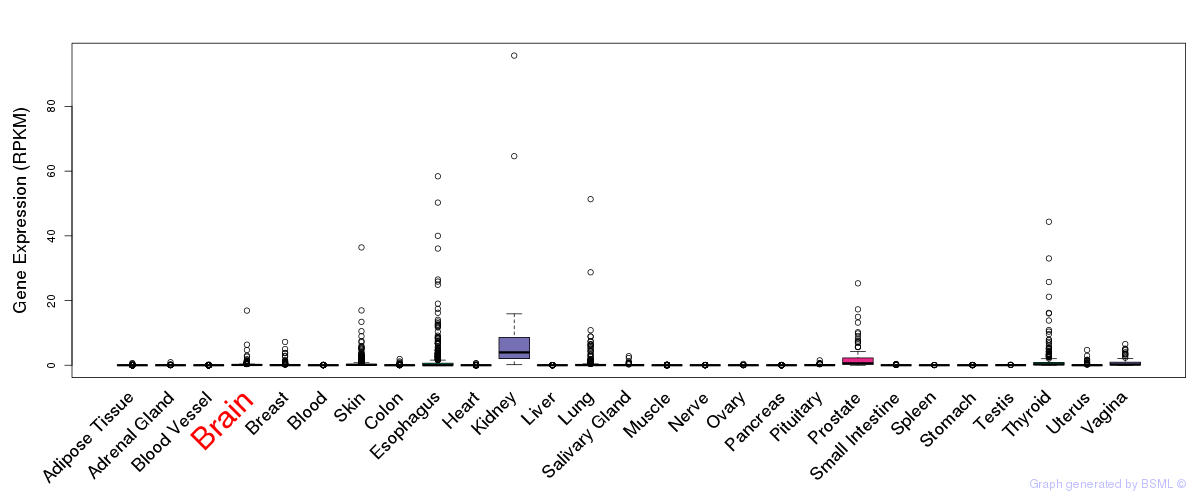
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| COX6B1 | 0.83 | 0.79 |
| CYB5A | 0.79 | 0.76 |
| ZDHHC4 | 0.78 | 0.77 |
| CSTB | 0.78 | 0.70 |
| HBXIP | 0.77 | 0.77 |
| WBP1 | 0.76 | 0.70 |
| MRPS24 | 0.75 | 0.75 |
| NDUFB4 | 0.75 | 0.77 |
| CHCHD2 | 0.75 | 0.75 |
| ATP5L | 0.75 | 0.73 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| KIF16B | -0.43 | -0.41 |
| BAT2D1 | -0.42 | -0.40 |
| AP000769.2 | -0.41 | -0.57 |
| C9orf131 | -0.40 | -0.50 |
| BDP1 | -0.39 | -0.36 |
| SYNE1 | -0.39 | -0.37 |
| MALAT1 | -0.39 | -0.52 |
| RYR3 | -0.38 | -0.41 |
| JAKMIP3 | -0.38 | -0.41 |
| HERC2P2 | -0.38 | -0.42 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | 35 | 23 | All SZGR 2.0 genes in this pathway |
| REACTOME BIOLOGICAL OXIDATIONS | 139 | 91 | All SZGR 2.0 genes in this pathway |
| REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | 51 | 34 | All SZGR 2.0 genes in this pathway |
| REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | 70 | 50 | All SZGR 2.0 genes in this pathway |
| REACTOME STEROID HORMONES | 29 | 21 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF LIPIDS AND LIPOPROTEINS | 478 | 302 | All SZGR 2.0 genes in this pathway |
| NAKAMURA TUMOR ZONE PERIPHERAL VS CENTRAL DN | 634 | 384 | All SZGR 2.0 genes in this pathway |
| KIM RESPONSE TO TSA AND DECITABINE UP | 129 | 73 | All SZGR 2.0 genes in this pathway |
| DEURIG T CELL PROLYMPHOCYTIC LEUKEMIA UP | 368 | 234 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC3 TARGETS DN | 536 | 332 | All SZGR 2.0 genes in this pathway |
| NAGASHIMA NRG1 SIGNALING UP | 176 | 123 | All SZGR 2.0 genes in this pathway |
| KIM WT1 TARGETS DN | 459 | 276 | All SZGR 2.0 genes in this pathway |
| KIM WT1 TARGETS 12HR DN | 209 | 122 | All SZGR 2.0 genes in this pathway |
| LINDGREN BLADDER CANCER CLUSTER 3 DN | 229 | 142 | All SZGR 2.0 genes in this pathway |
| CHIN BREAST CANCER COPY NUMBER UP | 27 | 18 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY SERUM DEPRIVATION DN | 234 | 147 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN DN | 1781 | 1082 | All SZGR 2.0 genes in this pathway |
| YANAGIHARA ESX1 TARGETS | 30 | 19 | All SZGR 2.0 genes in this pathway |
| BUYTAERT PHOTODYNAMIC THERAPY STRESS DN | 637 | 377 | All SZGR 2.0 genes in this pathway |
| RICKMAN HEAD AND NECK CANCER A | 100 | 63 | All SZGR 2.0 genes in this pathway |
| BENPORATH SUZ12 TARGETS | 1038 | 678 | All SZGR 2.0 genes in this pathway |
| BENPORATH EED TARGETS | 1062 | 725 | All SZGR 2.0 genes in this pathway |
| BENPORATH ES WITH H3K27ME3 | 1118 | 744 | All SZGR 2.0 genes in this pathway |
| BENPORATH PRC2 TARGETS | 652 | 441 | All SZGR 2.0 genes in this pathway |
| GEORGES TARGETS OF MIR192 AND MIR215 | 893 | 528 | All SZGR 2.0 genes in this pathway |
| NIKOLSKY BREAST CANCER 20Q12 Q13 AMPLICON | 149 | 76 | All SZGR 2.0 genes in this pathway |
| SHEPARD CRUSH AND BURN MUTANT UP | 197 | 110 | All SZGR 2.0 genes in this pathway |
| MATSUDA NATURAL KILLER DIFFERENTIATION | 475 | 313 | All SZGR 2.0 genes in this pathway |
| TAKAO RESPONSE TO UVB RADIATION DN | 98 | 67 | All SZGR 2.0 genes in this pathway |
| BILD CTNNB1 ONCOGENIC SIGNATURE | 82 | 52 | All SZGR 2.0 genes in this pathway |
| STEIN ESRRA TARGETS UP | 388 | 234 | All SZGR 2.0 genes in this pathway |
| OSADA ASCL1 TARGETS DN | 24 | 20 | All SZGR 2.0 genes in this pathway |
| BRUECKNER TARGETS OF MIRLET7A3 UP | 111 | 69 | All SZGR 2.0 genes in this pathway |
| MOOTHA HUMAN MITODB 6 2002 | 429 | 260 | All SZGR 2.0 genes in this pathway |
| TOOKER GEMCITABINE RESISTANCE DN | 122 | 84 | All SZGR 2.0 genes in this pathway |
| MEISSNER NPC HCP WITH H3 UNMETHYLATED | 536 | 296 | All SZGR 2.0 genes in this pathway |
| MEISSNER BRAIN HCP WITH H3K27ME3 | 269 | 159 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN MCV6 HCP WITH H3K27ME3 | 435 | 318 | All SZGR 2.0 genes in this pathway |
| STEIN ESR1 TARGETS | 85 | 55 | All SZGR 2.0 genes in this pathway |
| STEIN ESRRA TARGETS | 535 | 325 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN MEF HCP WITH H3K27ME3 | 590 | 403 | All SZGR 2.0 genes in this pathway |
| STAMBOLSKY RESPONSE TO VITAMIN D3 UP | 84 | 48 | All SZGR 2.0 genes in this pathway |
| LU EZH2 TARGETS DN | 414 | 237 | All SZGR 2.0 genes in this pathway |
| CERIBELLI PROMOTERS INACTIVE AND BOUND BY NFY | 44 | 20 | All SZGR 2.0 genes in this pathway |
| WINZEN DEGRADED VIA KHSRP | 100 | 70 | All SZGR 2.0 genes in this pathway |
| BOSCO INTERFERON INDUCED ANTIVIRAL MODULE | 78 | 48 | All SZGR 2.0 genes in this pathway |
| LIM MAMMARY LUMINAL PROGENITOR UP | 58 | 30 | All SZGR 2.0 genes in this pathway |