Gene Page: MDGA2
Summary ?
| GeneID | 161357 |
| Symbol | MDGA2 |
| Synonyms | MAMDC1|c14_5286 |
| Description | MAM domain containing glycosylphosphatidylinositol anchor 2 |
| Reference | MIM:611128|HGNC:HGNC:19835|Ensembl:ENSG00000139915|HPRD:14352|Vega:OTTHUMG00000029429 |
| Gene type | protein-coding |
| Map location | 14q21.3 |
| Pascal p-value | 4.236E-4 |
| Fetal beta | -0.246 |
| DMG | 2 (# studies) |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 2 |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 2 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg13687915 | 14 | 47311141 | MDGA2 | 3.696E-4 | -0.628 | 0.043 | DMG:Wockner_2014 |
| cg18115064 | 14 | 48143618 | MDGA2 | 4.58E-10 | -0.022 | 8.11E-7 | DMG:Jaffe_2016 |
Section II. Transcriptome annotation
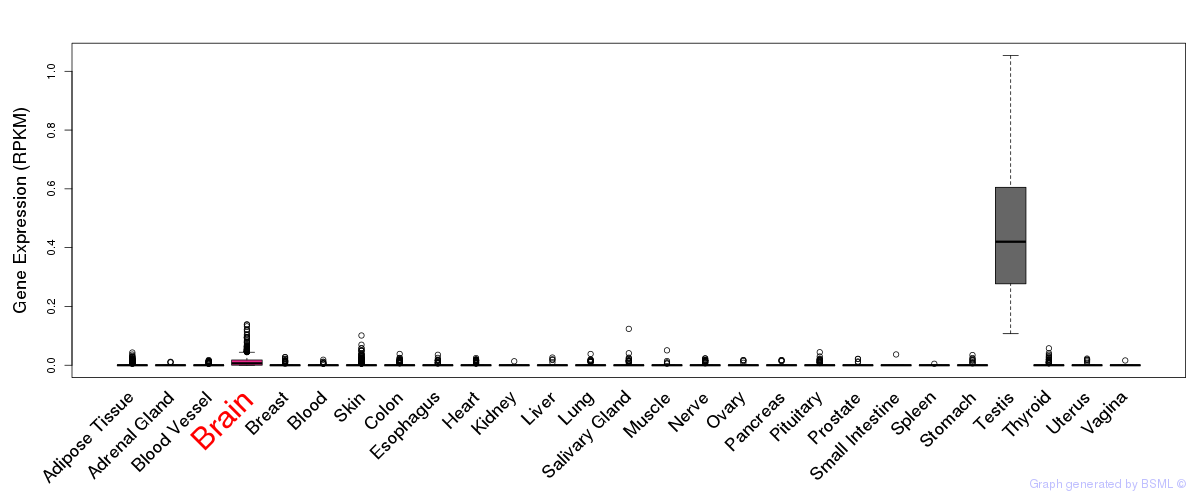
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0021522 | spinal cord motor neuron differentiation | ISS | neuron, Brain (GO term level: 9) | 15019943 |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005737 | cytoplasm | IDA | 18029348 | |
| GO:0005886 | plasma membrane | IEA | - | |
| GO:0031225 | anchored to membrane | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| TAKADA GASTRIC CANCER COPY NUMBER DN | 29 | 15 | All SZGR 2.0 genes in this pathway |
| DAWSON METHYLATED IN LYMPHOMA TCL1 | 59 | 45 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-10 | 437 | 444 | 1A,m8 | hsa-miR-10a | UACCCUGUAGAUCCGAAUUUGUG |
| hsa-miR-10b | UACCCUGUAGAACCGAAUUUGU | ||||
| miR-124/506 | 817 | 823 | m8 | hsa-miR-506 | UAAGGCACCCUUCUGAGUAGA |
| hsa-miR-124brain | UAAGGCACGCGGUGAAUGCC | ||||
| miR-129-5p | 1409 | 1415 | 1A | hsa-miR-129brain | CUUUUUGCGGUCUGGGCUUGC |
| hsa-miR-129-5p | CUUUUUGCGGUCUGGGCUUGCU | ||||
| miR-130/301 | 861 | 867 | m8 | hsa-miR-130abrain | CAGUGCAAUGUUAAAAGGGCAU |
| hsa-miR-301 | CAGUGCAAUAGUAUUGUCAAAGC | ||||
| hsa-miR-130bbrain | CAGUGCAAUGAUGAAAGGGCAU | ||||
| hsa-miR-454-3p | UAGUGCAAUAUUGCUUAUAGGGUUU | ||||
| miR-139 | 1752 | 1758 | 1A | hsa-miR-139brain | UCUACAGUGCACGUGUCU |
| miR-140 | 781 | 787 | m8 | hsa-miR-140brain | AGUGGUUUUACCCUAUGGUAG |
| miR-148/152 | 862 | 868 | m8 | hsa-miR-148a | UCAGUGCACUACAGAACUUUGU |
| hsa-miR-152brain | UCAGUGCAUGACAGAACUUGGG | ||||
| hsa-miR-148b | UCAGUGCAUCACAGAACUUUGU | ||||
| miR-153 | 1643 | 1649 | 1A | hsa-miR-153 | UUGCAUAGUCACAAAAGUGA |
| miR-200bc/429 | 1664 | 1670 | 1A | hsa-miR-200b | UAAUACUGCCUGGUAAUGAUGAC |
| hsa-miR-200c | UAAUACUGCCGGGUAAUGAUGG | ||||
| hsa-miR-429 | UAAUACUGUCUGGUAAAACCGU | ||||
| miR-21 | 634 | 640 | 1A | hsa-miR-21brain | UAGCUUAUCAGACUGAUGUUGA |
| hsa-miR-590 | GAGCUUAUUCAUAAAAGUGCAG | ||||
| miR-218 | 1700 | 1707 | 1A,m8 | hsa-miR-218brain | UUGUGCUUGAUCUAACCAUGU |
| hsa-miR-218brain | UUGUGCUUGAUCUAACCAUGU | ||||
| miR-299-5p | 687 | 693 | m8 | hsa-miR-299-5p | UGGUUUACCGUCCCACAUACAU |
| miR-30-3p | 1289 | 1295 | 1A | hsa-miR-30a-3p | CUUUCAGUCGGAUGUUUGCAGC |
| hsa-miR-30e-3p | CUUUCAGUCGGAUGUUUACAGC | ||||
| miR-33 | 1483 | 1489 | m8 | hsa-miR-33 | GUGCAUUGUAGUUGCAUUG |
| hsa-miR-33b | GUGCAUUGCUGUUGCAUUGCA | ||||
| miR-363 | 1766 | 1772 | 1A | hsa-miR-363 | AUUGCACGGUAUCCAUCUGUAA |
| hsa-miR-363 | AUUGCACGGUAUCCAUCUGUAA | ||||
| hsa-miR-363 | AUUGCACGGUAUCCAUCUGUAA | ||||
| miR-380-5p | 1120 | 1127 | 1A,m8 | hsa-miR-380-5p | UGGUUGACCAUAGAACAUGCGC |
| hsa-miR-563 | AGGUUGACAUACGUUUCCC | ||||
| miR-382 | 1783 | 1789 | 1A | hsa-miR-382brain | GAAGUUGUUCGUGGUGGAUUCG |
| miR-448 | 1643 | 1649 | 1A | hsa-miR-448 | UUGCAUAUGUAGGAUGUCCCAU |
| miR-490 | 1104 | 1111 | 1A,m8 | hsa-miR-490 | CAACCUGGAGGACUCCAUGCUG |
| miR-494 | 368 | 374 | 1A | hsa-miR-494brain | UGAAACAUACACGGGAAACCUCUU |
| miR-7 | 1100 | 1106 | 1A | hsa-miR-7SZ | UGGAAGACUAGUGAUUUUGUUG |
| miR-9 | 87 | 94 | 1A,m8 | hsa-miR-9SZ | UCUUUGGUUAUCUAGCUGUAUGA |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.