Gene Page: FAM134C
Summary ?
| GeneID | 162427 |
| Symbol | FAM134C |
| Synonyms | - |
| Description | family with sequence similarity 134 member C |
| Reference | MIM:616498|HGNC:HGNC:27258|Ensembl:ENSG00000141699|HPRD:14108|Vega:OTTHUMG00000180275 |
| Gene type | protein-coding |
| Map location | 17q21.2 |
| Pascal p-value | 0.053 |
| DMG | 1 (# studies) |
| eGene | Anterior cingulate cortex BA24 Caudate basal ganglia Meta |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 2 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg02697074 | 17 | 40761760 | FAM134C | 2.3E-9 | -0.013 | 1.75E-6 | DMG:Jaffe_2016 |
| cg04328701 | 17 | 40761179 | FAM134C | 6.11E-8 | -0.013 | 1.53E-5 | DMG:Jaffe_2016 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs36005199 | 17 | 40597555 | FAM134C | ENSG00000141699.6 | 3.3648E-6 | 0.01 | 163849 | gtex_brain_ba24 |
| rs34460267 | 17 | 40615865 | FAM134C | ENSG00000141699.6 | 1.5732E-6 | 0.01 | 145539 | gtex_brain_ba24 |
| rs1032070 | 17 | 40618251 | FAM134C | ENSG00000141699.6 | 1.5732E-6 | 0.01 | 143153 | gtex_brain_ba24 |
| rs34807589 | 17 | 40624656 | FAM134C | ENSG00000141699.6 | 1.56803E-6 | 0.01 | 136748 | gtex_brain_ba24 |
| rs2006141 | 17 | 40679718 | FAM134C | ENSG00000141699.6 | 6.25079E-7 | 0.01 | 81686 | gtex_brain_ba24 |
| rs4633765 | 17 | 40682341 | FAM134C | ENSG00000141699.6 | 6.20285E-7 | 0.01 | 79063 | gtex_brain_ba24 |
| rs4793099 | 17 | 40686040 | FAM134C | ENSG00000141699.6 | 6.05693E-7 | 0.01 | 75364 | gtex_brain_ba24 |
| rs2071046 | 17 | 40689613 | FAM134C | ENSG00000141699.6 | 4.89406E-7 | 0.01 | 71791 | gtex_brain_ba24 |
| rs4792923 | 17 | 40690118 | FAM134C | ENSG00000141699.6 | 4.50325E-7 | 0.01 | 71286 | gtex_brain_ba24 |
Section II. Transcriptome annotation
General gene expression (GTEx)
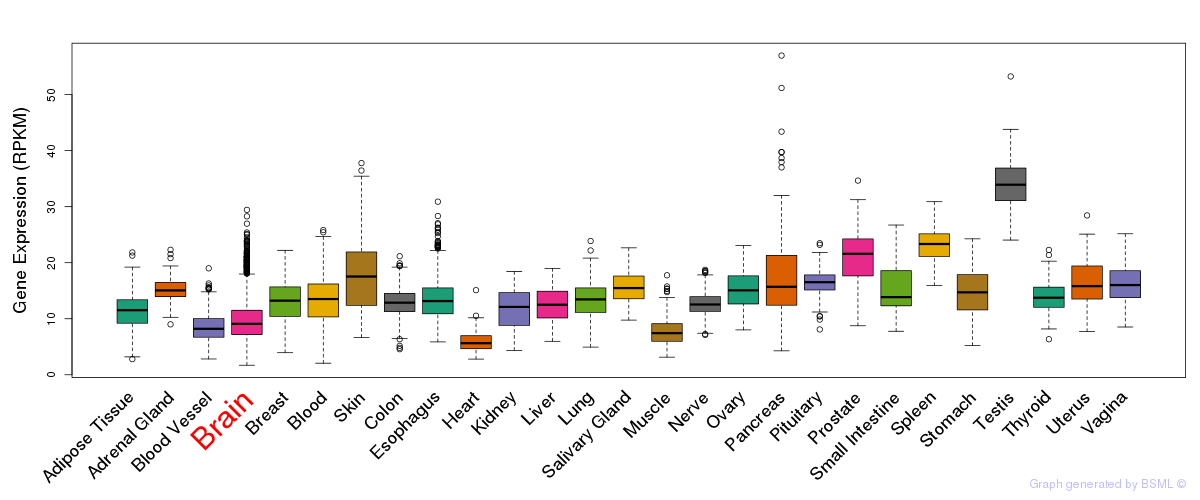
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| RODRIGUES THYROID CARCINOMA POORLY DIFFERENTIATED DN | 805 | 505 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY SERUM DEPRIVATION UP | 552 | 347 | All SZGR 2.0 genes in this pathway |
| PEREZ TP53 TARGETS | 1174 | 695 | All SZGR 2.0 genes in this pathway |
| TSAI RESPONSE TO IONIZING RADIATION | 149 | 101 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 48HR UP | 487 | 286 | All SZGR 2.0 genes in this pathway |
| BENPORATH NANOG TARGETS | 988 | 594 | All SZGR 2.0 genes in this pathway |
| BENPORATH SOX2 TARGETS | 734 | 436 | All SZGR 2.0 genes in this pathway |
| CHIARETTI ACUTE LYMPHOBLASTIC LEUKEMIA ZAP70 | 67 | 33 | All SZGR 2.0 genes in this pathway |
| DOUGLAS BMI1 TARGETS DN | 314 | 188 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 STIMULATED | 1022 | 619 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 UNSTIMULATED | 1229 | 713 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 TARGETS DN | 543 | 317 | All SZGR 2.0 genes in this pathway |
| MARTINEZ TP53 TARGETS DN | 593 | 372 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 AND TP53 TARGETS DN | 591 | 366 | All SZGR 2.0 genes in this pathway |
| RIZKI TUMOR INVASIVENESS 3D UP | 210 | 124 | All SZGR 2.0 genes in this pathway |
| GRADE COLON VS RECTAL CANCER DN | 56 | 36 | All SZGR 2.0 genes in this pathway |
| LEE LIVER CANCER SURVIVAL UP | 185 | 112 | All SZGR 2.0 genes in this pathway |
| AGUIRRE PANCREATIC CANCER COPY NUMBER UP | 298 | 174 | All SZGR 2.0 genes in this pathway |
| JOHNSTONE PARVB TARGETS 3 UP | 430 | 288 | All SZGR 2.0 genes in this pathway |
| GOBERT OLIGODENDROCYTE DIFFERENTIATION DN | 1080 | 713 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 1ST EGF PULSE ONLY | 1839 | 928 | All SZGR 2.0 genes in this pathway |
| ZWANG EGF INTERVAL DN | 214 | 124 | All SZGR 2.0 genes in this pathway |