Gene Page: ACE
Summary ?
| GeneID | 1636 |
| Symbol | ACE |
| Synonyms | ACE1|CD143|DCP|DCP1|ICH|MVCD3 |
| Description | angiotensin I converting enzyme |
| Reference | MIM:106180|HGNC:HGNC:2707|Ensembl:ENSG00000159640|HPRD:00108|Vega:OTTHUMG00000154927 |
| Gene type | protein-coding |
| Map location | 17q23.3 |
| Pascal p-value | 0.004 |
| Fetal beta | -0.53 |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenics,schizophrenias,schizotypal | Click to show details |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.089 |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
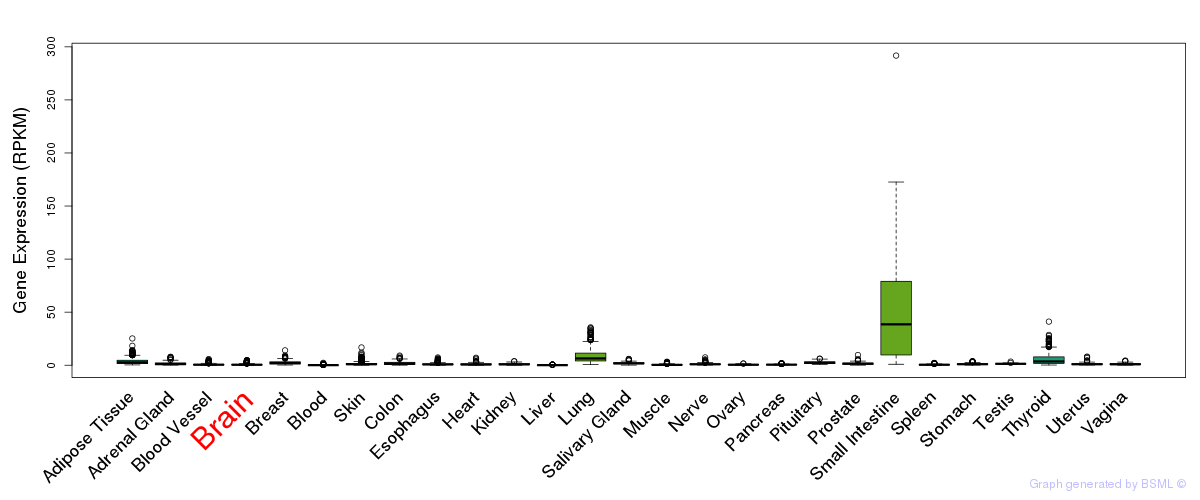
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0004180 | carboxypeptidase activity | IEA | - | |
| GO:0008270 | zinc ion binding | IEA | - | |
| GO:0008241 | peptidyl-dipeptidase activity | IDA | 1320019 |1668266 |2849100 |17077303 | |
| GO:0008241 | peptidyl-dipeptidase activity | IEA | - | |
| GO:0008241 | peptidyl-dipeptidase activity | ISS | 11303049 | |
| GO:0008237 | metallopeptidase activity | IDA | 1320019 |1668266 |2849100 |17077303 | |
| GO:0008237 | metallopeptidase activity | IEA | - | |
| GO:0008237 | metallopeptidase activity | ISS | - | |
| GO:0008144 | drug binding | IDA | 1320019 | |
| GO:0008233 | peptidase activity | IEA | - | |
| GO:0031404 | chloride ion binding | IDA | 12540854 | |
| GO:0031711 | bradykinin receptor binding | IPI | 17077303 | |
| GO:0046872 | metal ion binding | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0003081 | regulation of systemic arterial blood pressure by renin-angiotensin | IC | 1668266 | |
| GO:0002005 | angiotensin catabolic process in blood | IC | 1668266 | |
| GO:0001822 | kidney development | IMP | 16116425 | |
| GO:0006508 | proteolysis | IEA | - | |
| GO:0008217 | regulation of blood pressure | ISS | 11303049 | |
| GO:0042312 | regulation of vasodilation | IC | 4322742 | |
| GO:0014910 | regulation of smooth muscle cell migration | ISS | - | |
| GO:0042447 | hormone catabolic process | IDA | 7876104 | |
| GO:0042447 | hormone catabolic process | ISS | 11303049 | |
| GO:0019229 | regulation of vasoconstriction | IC | 1668266 | |
| GO:0032943 | mononuclear cell proliferation | IC | 7876104 | |
| GO:0043171 | peptide catabolic process | IDA | 4322742 | |
| GO:0050482 | arachidonic acid secretion | IDA | 17077303 | |
| GO:0060218 | hemopoietic stem cell differentiation | IC | 7876104 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005576 | extracellular region | IEA | - | |
| GO:0005615 | extracellular space | IDA | 1668266 |4322742 | |
| GO:0005624 | membrane fraction | IDA | 1668266 |2849100 | |
| GO:0005768 | endosome | IDA | 17077303 | |
| GO:0016020 | membrane | IEA | - | |
| GO:0016021 | integral to membrane | IEA | - | |
| GO:0005886 | plasma membrane | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG RENIN ANGIOTENSIN SYSTEM | 17 | 10 | All SZGR 2.0 genes in this pathway |
| KEGG HYPERTROPHIC CARDIOMYOPATHY HCM | 85 | 65 | All SZGR 2.0 genes in this pathway |
| BIOCARTA ACE2 PATHWAY | 13 | 7 | All SZGR 2.0 genes in this pathway |
| SA MMP CYTOKINE CONNECTION | 15 | 9 | All SZGR 2.0 genes in this pathway |
| GINESTIER BREAST CANCER 20Q13 AMPLIFICATION DN | 180 | 101 | All SZGR 2.0 genes in this pathway |
| TAKEDA TARGETS OF NUP98 HOXA9 FUSION 10D DN | 142 | 90 | All SZGR 2.0 genes in this pathway |
| CHIARADONNA NEOPLASTIC TRANSFORMATION CDC25 DN | 153 | 100 | All SZGR 2.0 genes in this pathway |
| BENPORATH MYC TARGETS WITH EBOX | 230 | 156 | All SZGR 2.0 genes in this pathway |
| SANA TNF SIGNALING DN | 90 | 57 | All SZGR 2.0 genes in this pathway |
| BYSTRYKH HEMATOPOIESIS STEM CELL QTL TRANS | 882 | 572 | All SZGR 2.0 genes in this pathway |
| SANSOM APC TARGETS DN | 366 | 238 | All SZGR 2.0 genes in this pathway |
| ABE VEGFA TARGETS | 20 | 15 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS STEM CELL AND PROGENITOR | 681 | 420 | All SZGR 2.0 genes in this pathway |
| LEIN CHOROID PLEXUS MARKERS | 103 | 61 | All SZGR 2.0 genes in this pathway |
| KYNG DNA DAMAGE BY UV | 62 | 43 | All SZGR 2.0 genes in this pathway |
| KYNG DNA DAMAGE DN | 195 | 135 | All SZGR 2.0 genes in this pathway |
| MATZUK SPERMATOZOA | 114 | 77 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS UP | 1305 | 895 | All SZGR 2.0 genes in this pathway |
| SWEET LUNG CANCER KRAS DN | 435 | 289 | All SZGR 2.0 genes in this pathway |
| CHEN METABOLIC SYNDROM NETWORK | 1210 | 725 | All SZGR 2.0 genes in this pathway |
| MEISSNER BRAIN HCP WITH H3K4ME3 AND H3K27ME3 | 1069 | 729 | All SZGR 2.0 genes in this pathway |
| ROME INSULIN TARGETS IN MUSCLE DN | 204 | 114 | All SZGR 2.0 genes in this pathway |
| MEISSNER NPC HCP WITH H3K4ME2 | 491 | 319 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN MCV6 HCP WITH H3K27ME3 | 435 | 318 | All SZGR 2.0 genes in this pathway |
| DANG BOUND BY MYC | 1103 | 714 | All SZGR 2.0 genes in this pathway |
| MARTENS TRETINOIN RESPONSE UP | 857 | 456 | All SZGR 2.0 genes in this pathway |
| FEVR CTNNB1 TARGETS UP | 682 | 433 | All SZGR 2.0 genes in this pathway |
| PEDRIOLI MIR31 TARGETS DN | 418 | 245 | All SZGR 2.0 genes in this pathway |
| ACEVEDO FGFR1 TARGETS IN PROSTATE CANCER MODEL DN | 308 | 187 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-27 | 222 | 228 | 1A | hsa-miR-27abrain | UUCACAGUGGCUAAGUUCCGC |
| hsa-miR-27bbrain | UUCACAGUGGCUAAGUUCUGC | ||||
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.