Gene Page: AP1G1
Summary ?
| GeneID | 164 |
| Symbol | AP1G1 |
| Synonyms | ADTG|CLAPG1 |
| Description | adaptor related protein complex 1 gamma 1 subunit |
| Reference | MIM:603533|HGNC:HGNC:555|Ensembl:ENSG00000166747|HPRD:04637|Vega:OTTHUMG00000176871 |
| Gene type | protein-coding |
| Map location | 16q23 |
| Pascal p-value | 0.059 |
| Sherlock p-value | 0.277 |
| Fetal beta | 0.215 |
| DMG | 1 (# studies) |
| eGene | Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg16666698 | 16 | 71842293 | AP1G1 | 3.1E-8 | -0.013 | 9.43E-6 | DMG:Jaffe_2016 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs17000249 | chr4 | 76031156 | AP1G1 | 164 | 0.08 | trans | ||
| rs3217906 | chr12 | 4405803 | AP1G1 | 164 | 0.1 | trans |
Section II. Transcriptome annotation
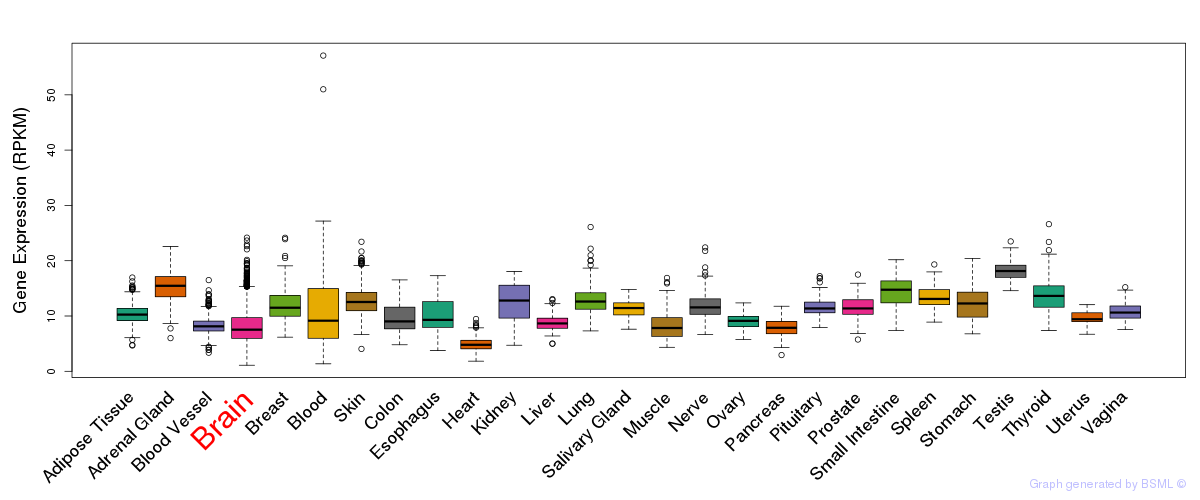
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| AFTPH | FLJ20080 | FLJ23793 | MGC33965 | Nbla10388 | aftiphilin | - | HPRD | 14665628 |
| AP1B1 | ADTB1 | AP105A | BAM22 | CLAPB2 | adaptor-related protein complex 1, beta 1 subunit | - | HPRD,BioGRID | 9733768 |
| AP1G1 | ADTG | CLAPG1 | MGC18255 | adaptor-related protein complex 1, gamma 1 subunit | - | HPRD | 12042876 |
| AP1GBP1 | MGC104959 | SYNG | AP1 gamma subunit binding protein 1 | - | HPRD,BioGRID | 9653655 |10814529 |12042876 |
| AP1M1 | AP47 | CLAPM2 | CLTNM | MU-1A | adaptor-related protein complex 1, mu 1 subunit | Affinity Capture-Western | BioGRID | 10535737 |
| AP1M2 | AP1-mu2 | HSMU1B | MU-1B | MU1B | mu2 | adaptor-related protein complex 1, mu 2 subunit | - | HPRD,BioGRID | 10535737 |
| AP1S1 | AP19 | CLAPS1 | FLJ92436 | SIGMA1A | WUGSC:H_DJ0747G18.2 | adaptor-related protein complex 1, sigma 1 subunit | The gamma-1 and sigma-1A subunits of the AP-1 complex interact. This interaction was modeled on a demonstrated interaction between mouse gamma-1 and human sigma-1A. | BIND | 15469849 |
| AP1S1 | AP19 | CLAPS1 | FLJ92436 | SIGMA1A | WUGSC:H_DJ0747G18.2 | adaptor-related protein complex 1, sigma 1 subunit | - | HPRD,BioGRID | 9733768 |
| AP1S2 | DC22 | MGC:1902 | MRX59 | SIGMA1B | adaptor-related protein complex 1, sigma 2 subunit | - | HPRD,BioGRID | 9733768 |
| ARF1 | - | ADP-ribosylation factor 1 | - | HPRD,BioGRID | 11926829 |
| C4orf16 | 2C18 | GBAR | PRO0971 | chromosome 4 open reading frame 16 | C4orf16 (2c18) interacts with AP1G1 (gamma-1-adaptin). | BIND | 15775984 |
| CLINT1 | CLINT | ENTH | EPN4 | EPNR | EPSINR | FLJ46753 | KIAA0171 | clathrin interactor 1 | - | HPRD,BioGRID | 12429846 |
| CLTC | CHC | CHC17 | CLH-17 | CLTCL2 | Hc | KIAA0034 | clathrin, heavy chain (Hc) | - | HPRD,BioGRID | 11451993 |
| DNER | UNQ26 | bet | delta/notch-like EGF repeat containing | - | HPRD | 11950833 |
| KIF13A | FLJ27232 | bA500C11.2 | kinesin family member 13A | - | HPRD,BioGRID | 11106728 |
| MAP1A | FLJ77111 | MAP1L | MTAP1A | microtubule-associated protein 1A | - | HPRD | 10747088 |11418592 |
| NECAP1 | DKFZP566B183 | MGC131900 | NECAP endocytosis associated 1 | Affinity Capture-Western | BioGRID | 14665628 |
| NECAP2 | FLJ10420 | RP4-798A10.1 | NECAP endocytosis associated 2 | Affinity Capture-Western | BioGRID | 14665628 |
| RAB5A | RAB5 | RAB5A, member RAS oncogene family | Two-hybrid | BioGRID | 14665628 |
| RABEP1 | RAB5EP | RABPT5 | rabaptin, RAB GTPase binding effector protein 1 | Reconstituted Complex Two-hybrid | BioGRID | 12042876 |12505986 |
| SPTB | HSpTB1 | spectrin, beta, erythrocytic | - | HPRD | 10477754 |
| SYP | - | synaptophysin | Two-hybrid | BioGRID | 12498786 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG LYSOSOME | 121 | 83 | All SZGR 2.0 genes in this pathway |
| REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | 10 | 5 | All SZGR 2.0 genes in this pathway |
| REACTOME MEMBRANE TRAFFICKING | 129 | 74 | All SZGR 2.0 genes in this pathway |
| REACTOME NEF MEDIATES DOWN MODULATION OF CELL SURFACE RECEPTORS BY RECRUITING THEM TO CLATHRIN ADAPTERS | 21 | 12 | All SZGR 2.0 genes in this pathway |
| REACTOME TRANS GOLGI NETWORK VESICLE BUDDING | 60 | 31 | All SZGR 2.0 genes in this pathway |
| REACTOME MHC CLASS II ANTIGEN PRESENTATION | 91 | 61 | All SZGR 2.0 genes in this pathway |
| REACTOME LYSOSOME VESICLE BIOGENESIS | 23 | 11 | All SZGR 2.0 genes in this pathway |
| REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | 53 | 27 | All SZGR 2.0 genes in this pathway |
| REACTOME HIV INFECTION | 207 | 122 | All SZGR 2.0 genes in this pathway |
| REACTOME HOST INTERACTIONS OF HIV FACTORS | 132 | 81 | All SZGR 2.0 genes in this pathway |
| REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | 28 | 19 | All SZGR 2.0 genes in this pathway |
| REACTOME IMMUNE SYSTEM | 933 | 616 | All SZGR 2.0 genes in this pathway |
| REACTOME ADAPTIVE IMMUNE SYSTEM | 539 | 350 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN DN | 1781 | 1082 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN RESPONSE TO MC AND DOXORUBICIN DN | 770 | 415 | All SZGR 2.0 genes in this pathway |
| MARTORIATI MDM4 TARGETS FETAL LIVER DN | 514 | 319 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA1 PCC NETWORK | 1652 | 1023 | All SZGR 2.0 genes in this pathway |
| BENPORATH NANOG TARGETS | 988 | 594 | All SZGR 2.0 genes in this pathway |
| BYSTROEM CORRELATED WITH IL5 DN | 64 | 47 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS EARLY PROGENITOR | 532 | 309 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 5 | 482 | 296 | All SZGR 2.0 genes in this pathway |
| KYNG DNA DAMAGE BY GAMMA RADIATION | 81 | 59 | All SZGR 2.0 genes in this pathway |
| KYNG DNA DAMAGE UP | 226 | 164 | All SZGR 2.0 genes in this pathway |
| HAN SATB1 TARGETS UP | 395 | 249 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS DN | 1972 | 1213 | All SZGR 2.0 genes in this pathway |
| JOHNSTONE PARVB TARGETS 3 DN | 918 | 550 | All SZGR 2.0 genes in this pathway |