Gene Page: DMD
Summary ?
| GeneID | 1756 |
| Symbol | DMD |
| Synonyms | BMD|CMD3B|DXS142|DXS164|DXS206|DXS230|DXS239|DXS268|DXS269|DXS270|DXS272|MRX85 |
| Description | dystrophin |
| Reference | MIM:300377|HGNC:HGNC:2928|Ensembl:ENSG00000198947|HPRD:02303|Vega:OTTHUMG00000021336 |
| Gene type | protein-coding |
| Map location | Xp21.2 |
| Sherlock p-value | 0.875 |
| Fetal beta | -0.3 |
| Support | CompositeSet |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWAScat | Genome-wide Association Studies | This data set includes 560 SNPs associated with schizophrenia. A total of 486 genes were mapped to these SNPs within 50kb. | |
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DNM:Fromer_2014 | Whole Exome Sequencing analysis | This study reported a WES study of 623 schizophrenia trios, reporting DNMs using genomic DNA. | |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 2 | |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.259 |
Section I. Genetics and epigenetics annotation
 DNM table
DNM table
| Gene | Chromosome | Position | Ref | Alt | Transcript | AA change | Mutation type | Sift | CG46 | Trait | Study |
|---|---|---|---|---|---|---|---|---|---|---|---|
| DMD | chrX | 32716053 | G | C | NM_000109 NM_004006 NM_004007 NM_004009 NM_004010 | p.290S>R p.298S>R p.175S>R p.294S>R p.175S>R | missense missense missense missense missense | Schizophrenia | DNM:Fromer_2014 |
Section II. Transcriptome annotation
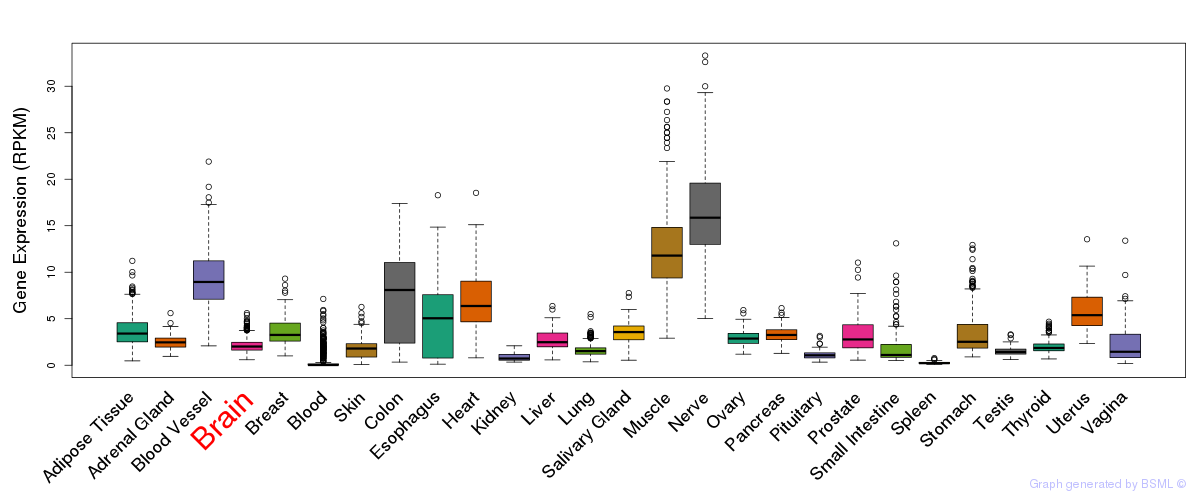
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| C10orf79 | 0.93 | 0.62 |
| C12orf63 | 0.93 | 0.54 |
| WDR52 | 0.93 | 0.57 |
| DNAH3 | 0.92 | 0.38 |
| CCDC40 | 0.92 | 0.60 |
| SPAG17 | 0.92 | 0.59 |
| C3orf15 | 0.91 | 0.71 |
| DNAH12 | 0.91 | 0.58 |
| DNAH2 | 0.91 | 0.75 |
| LRP2BP | 0.89 | 0.42 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| MT-CO2 | -0.23 | -0.40 |
| AF347015.31 | -0.23 | -0.43 |
| COX4I2 | -0.22 | -0.40 |
| MYL3 | -0.22 | -0.43 |
| AF347015.8 | -0.22 | -0.38 |
| AF347015.27 | -0.22 | -0.37 |
| AF347015.2 | -0.22 | -0.37 |
| AF347015.33 | -0.21 | -0.35 |
| CXCL14 | -0.21 | -0.37 |
| MUSTN1 | -0.21 | -0.34 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0003779 | actin binding | IEA | - | |
| GO:0003779 | actin binding | ISS | 2261642 | |
| GO:0003779 | actin binding | TAS | 12376554 | |
| GO:0005509 | calcium ion binding | IEA | - | |
| GO:0005515 | protein binding | IEA | - | |
| GO:0005515 | protein binding | IPI | 16000376 | |
| GO:0005200 | structural constituent of cytoskeleton | TAS | 3282674 | |
| GO:0008270 | zinc ion binding | IEA | - | |
| GO:0008307 | structural constituent of muscle | IDA | 16000376 | |
| GO:0008307 | structural constituent of muscle | TAS | 3287171 | |
| GO:0050998 | nitric-oxide synthase binding | ISS | 7545544 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007519 | skeletal muscle development | IEA | neuron (GO term level: 8) | - |
| GO:0045213 | neurotransmitter receptor metabolic process | IEA | Synap, Neurotransmitter (GO term level: 7) | - |
| GO:0007517 | muscle development | NAS | 1824797 | |
| GO:0007016 | cytoskeletal anchoring at plasma membrane | ISS | 2261642 | |
| GO:0007016 | cytoskeletal anchoring at plasma membrane | NAS | - | |
| GO:0043043 | peptide biosynthetic process | IDA | 16000376 | |
| GO:0046716 | muscle maintenance | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0045202 | synapse | IEA | neuron, Synap, Neurotransmitter, Glial (GO term level: 2) | - |
| GO:0005792 | microsome | IEA | - | |
| GO:0005856 | cytoskeleton | TAS | 3282674 | |
| GO:0005626 | insoluble fraction | IEA | - | |
| GO:0005634 | nucleus | IDA | 18029348 | |
| GO:0005737 | cytoplasm | IEA | - | |
| GO:0030018 | Z disc | IEA | - | |
| GO:0009986 | cell surface | IDA | 10867799 | |
| GO:0016010 | dystrophin-associated glycoprotein complex | IEA | - | |
| GO:0016010 | dystrophin-associated glycoprotein complex | ISS | 2261642 | |
| GO:0016010 | dystrophin-associated glycoprotein complex | NAS | - | |
| GO:0016010 | dystrophin-associated glycoprotein complex | TAS | 8282811 | |
| GO:0042383 | sarcolemma | IDA | 7545544 | |
| GO:0042383 | sarcolemma | IEA | - | |
| GO:0030055 | cell-substrate junction | IEA | - | |
| GO:0043034 | costamere | IDA | 16000376 | |
| GO:0045121 | membrane raft | IEA | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ACTA1 | ACTA | ASMA | CFTD | CFTD1 | CFTDM | MPFD | NEM1 | NEM2 | NEM3 | actin, alpha 1, skeletal muscle | - | HPRD,BioGRID | 1544421|11259305 |
| ACTA1 | ACTA | ASMA | CFTD | CFTD1 | CFTDM | MPFD | NEM1 | NEM2 | NEM3 | actin, alpha 1, skeletal muscle | - | HPRD | 1544421 |
| ACTC1 | ACTC | CMD1R | CMH11 | actin, alpha, cardiac muscle 1 | - | HPRD | 11997265 |
| CADPS | CADPS1 | CAPS | CAPS1 | KIAA1121 | Ca++-dependent secretion activator | - | HPRD | 12659812 |
| CADPS2 | FLJ40851 | KIAA1591 | Ca++-dependent secretion activator 2 | - | HPRD | 12659812 |
| DAG1 | 156DAG | A3a | AGRNR | DAG | dystroglycan 1 (dystrophin-associated glycoprotein 1) | - | HPRD,BioGRID | 10355629 |10932245 |
| DGKZ | DAGK5 | DAGK6 | DGK-ZETA | hDGKzeta | diacylglycerol kinase, zeta 104kDa | - | HPRD | 11352924 |
| DTNA | D18S892E | DRP3 | DTN | FLJ96209 | LVNC1 | dystrobrevin, alpha | - | HPRD,BioGRID | 9356463 |
| DTNB | MGC17163 | MGC57126 | dystrobrevin, beta | Affinity Capture-Western | BioGRID | 10545507 |
| FASLG | APT1LG1 | CD178 | CD95L | FASL | TNFSF6 | Fas ligand (TNF superfamily, member 6) | - | HPRD | 11741599 |
| KCNJ12 | FLJ14167 | IRK2 | KCNJN1 | Kir2.2 | Kir2.2v | hIRK | hIRK1 | hkir2.2x | kcnj12x | potassium inwardly-rectifying channel, subfamily J, member 12 | - | HPRD,BioGRID | 15024025 |
| KCNJ4 | HIR | HIRK2 | HRK1 | IRK3 | Kir2.3 | MGC142066 | MGC142068 | potassium inwardly-rectifying channel, subfamily J, member 4 | Affinity Capture-Western | BioGRID | 15024025 |
| PGM5 | PGMRP | phosphoglucomutase 5 | - | HPRD | 7890770 |10867799 |
| SGCZ | MGC149397 | ZSG1 | sarcoglycan zeta | - | HPRD | 12189167 |
| SNTA1 | SNT1 | TACIP1 | dJ1187J4.5 | syntrophin, alpha 1 (dystrophin-associated protein A1, 59kDa, acidic component) | - | HPRD,BioGRID | 7890602 |
| SNTB1 | 59-DAP | A1B | BSYN2 | DAPA1B | FLJ22442 | MGC111389 | SNT2 | SNT2B1 | TIP-43 | syntrophin, beta 1 (dystrophin-associated protein A1, 59kDa, basic component 1) | - | HPRD,BioGRID | 7844150 |
| SNTB2 | D16S2531E | EST25263 | SNT2B2 | SNT3 | SNTL | syntrophin, beta 2 (dystrophin-associated protein A1, 59kDa, basic component 2) | Protein-peptide | BioGRID | 8576247 |
| SNTG1 | G1SYN | SYN4 | syntrophin, gamma 1 | - | HPRD | 10747910 |11352924 |
| SNTG2 | G2SYN | MGC133174 | SYN5 | syntrophin, gamma 2 | - | HPRD | 10747910 |
| UTRN | DMDL | DRP | DRP1 | FLJ23678 | utrophin | - | HPRD | 10767429 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG HYPERTROPHIC CARDIOMYOPATHY HCM | 85 | 65 | All SZGR 2.0 genes in this pathway |
| KEGG ARRHYTHMOGENIC RIGHT VENTRICULAR CARDIOMYOPATHY ARVC | 76 | 59 | All SZGR 2.0 genes in this pathway |
| KEGG DILATED CARDIOMYOPATHY | 92 | 68 | All SZGR 2.0 genes in this pathway |
| KEGG VIRAL MYOCARDITIS | 73 | 58 | All SZGR 2.0 genes in this pathway |
| BIOCARTA AGR PATHWAY | 36 | 31 | All SZGR 2.0 genes in this pathway |
| REACTOME STRIATED MUSCLE CONTRACTION | 27 | 12 | All SZGR 2.0 genes in this pathway |
| REACTOME MUSCLE CONTRACTION | 48 | 24 | All SZGR 2.0 genes in this pathway |
| NAKAMURA TUMOR ZONE PERIPHERAL VS CENTRAL DN | 634 | 384 | All SZGR 2.0 genes in this pathway |
| LIU PROSTATE CANCER DN | 481 | 290 | All SZGR 2.0 genes in this pathway |
| ONKEN UVEAL MELANOMA DN | 526 | 357 | All SZGR 2.0 genes in this pathway |
| SENGUPTA NASOPHARYNGEAL CARCINOMA WITH LMP1 UP | 408 | 247 | All SZGR 2.0 genes in this pathway |
| TURASHVILI BREAST LOBULAR CARCINOMA VS DUCTAL NORMAL DN | 91 | 53 | All SZGR 2.0 genes in this pathway |
| TURASHVILI BREAST LOBULAR CARCINOMA VS LOBULAR NORMAL UP | 94 | 59 | All SZGR 2.0 genes in this pathway |
| CORRE MULTIPLE MYELOMA UP | 74 | 45 | All SZGR 2.0 genes in this pathway |
| HUTTMANN B CLL POOR SURVIVAL UP | 276 | 187 | All SZGR 2.0 genes in this pathway |
| CHARAFE BREAST CANCER LUMINAL VS BASAL DN | 455 | 304 | All SZGR 2.0 genes in this pathway |
| CHARAFE BREAST CANCER LUMINAL VS MESENCHYMAL DN | 460 | 312 | All SZGR 2.0 genes in this pathway |
| WANG LMO4 TARGETS DN | 352 | 225 | All SZGR 2.0 genes in this pathway |
| VECCHI GASTRIC CANCER EARLY DN | 367 | 220 | All SZGR 2.0 genes in this pathway |
| GINESTIER BREAST CANCER 20Q13 AMPLIFICATION DN | 180 | 101 | All SZGR 2.0 genes in this pathway |
| TAKEDA TARGETS OF NUP98 HOXA9 FUSION 10D UP | 194 | 122 | All SZGR 2.0 genes in this pathway |
| BILBAN B CLL LPL UP | 63 | 39 | All SZGR 2.0 genes in this pathway |
| LEE NEURAL CREST STEM CELL DN | 118 | 79 | All SZGR 2.0 genes in this pathway |
| GRAHAM CML DIVIDING VS NORMAL QUIESCENT DN | 95 | 57 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA DN | 1375 | 806 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA POORLY DIFFERENTIATED DN | 805 | 505 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA DN | 77 | 52 | All SZGR 2.0 genes in this pathway |
| DELYS THYROID CANCER UP | 443 | 294 | All SZGR 2.0 genes in this pathway |
| BEGUM TARGETS OF PAX3 FOXO1 FUSION UP | 60 | 45 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 TTD DN | 84 | 63 | All SZGR 2.0 genes in this pathway |
| FARMER BREAST CANCER APOCRINE VS BASAL | 330 | 217 | All SZGR 2.0 genes in this pathway |
| FARMER BREAST CANCER BASAL VS LULMINAL | 330 | 215 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 DN | 855 | 609 | All SZGR 2.0 genes in this pathway |
| WEINMANN ADAPTATION TO HYPOXIA UP | 29 | 24 | All SZGR 2.0 genes in this pathway |
| WEINMANN ADAPTATION TO HYPOXIA DN | 41 | 33 | All SZGR 2.0 genes in this pathway |
| GRUETZMANN PANCREATIC CANCER DN | 203 | 134 | All SZGR 2.0 genes in this pathway |
| RICKMAN HEAD AND NECK CANCER A | 100 | 63 | All SZGR 2.0 genes in this pathway |
| ONDER CDH1 TARGETS 2 DN | 464 | 276 | All SZGR 2.0 genes in this pathway |
| IIZUKA LIVER CANCER PROGRESSION L0 L1 UP | 17 | 12 | All SZGR 2.0 genes in this pathway |
| TARTE PLASMA CELL VS PLASMABLAST UP | 398 | 262 | All SZGR 2.0 genes in this pathway |
| HASLINGER B CLL WITH MUTATED VH GENES | 18 | 14 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE DN | 1237 | 837 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| CHIBA RESPONSE TO TSA DN | 23 | 14 | All SZGR 2.0 genes in this pathway |
| KRASNOSELSKAYA ILF3 TARGETS DN | 46 | 38 | All SZGR 2.0 genes in this pathway |
| LEIN CEREBELLUM MARKERS | 85 | 47 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 TARGETS UP | 673 | 430 | All SZGR 2.0 genes in this pathway |
| MARTINEZ TP53 TARGETS DN | 593 | 372 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 AND TP53 TARGETS DN | 591 | 366 | All SZGR 2.0 genes in this pathway |
| RIZKI TUMOR INVASIVENESS 3D UP | 210 | 124 | All SZGR 2.0 genes in this pathway |
| RIGGI EWING SARCOMA PROGENITOR UP | 430 | 288 | All SZGR 2.0 genes in this pathway |
| MASRI RESISTANCE TO TAMOXIFEN AND AROMATASE INHIBITORS UP | 20 | 10 | All SZGR 2.0 genes in this pathway |
| ACEVEDO METHYLATED IN LIVER CANCER DN | 940 | 425 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER LUMINAL B DN | 564 | 326 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER NORMAL LIKE UP | 476 | 285 | All SZGR 2.0 genes in this pathway |
| HELLEBREKERS SILENCED DURING TUMOR ANGIOGENESIS | 80 | 56 | All SZGR 2.0 genes in this pathway |
| SPIRA SMOKERS LUNG CANCER UP | 38 | 24 | All SZGR 2.0 genes in this pathway |
| RUIZ TNC TARGETS DN | 142 | 79 | All SZGR 2.0 genes in this pathway |
| BOYAULT LIVER CANCER SUBCLASS G123 DN | 51 | 30 | All SZGR 2.0 genes in this pathway |
| YAGI AML WITH INV 16 TRANSLOCATION | 422 | 277 | All SZGR 2.0 genes in this pathway |
| KORKOLA EMBRYONIC CARCINOMA VS SEMINOMA DN | 25 | 14 | All SZGR 2.0 genes in this pathway |
| WONG ADULT TISSUE STEM MODULE | 721 | 492 | All SZGR 2.0 genes in this pathway |
| HIRSCH CELLULAR TRANSFORMATION SIGNATURE DN | 103 | 67 | All SZGR 2.0 genes in this pathway |
| GABRIELY MIR21 TARGETS | 289 | 187 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE VIA TP53 GROUP D | 280 | 158 | All SZGR 2.0 genes in this pathway |
| FEVR CTNNB1 TARGETS DN | 553 | 343 | All SZGR 2.0 genes in this pathway |
| IKEDA MIR30 TARGETS UP | 116 | 87 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| let-7/98 | 1848 | 1855 | 1A,m8 | hsa-let-7abrain | UGAGGUAGUAGGUUGUAUAGUU |
| hsa-let-7bbrain | UGAGGUAGUAGGUUGUGUGGUU | ||||
| hsa-let-7cbrain | UGAGGUAGUAGGUUGUAUGGUU | ||||
| hsa-let-7dbrain | AGAGGUAGUAGGUUGCAUAGU | ||||
| hsa-let-7ebrain | UGAGGUAGGAGGUUGUAUAGU | ||||
| hsa-let-7fbrain | UGAGGUAGUAGAUUGUAUAGUU | ||||
| hsa-miR-98brain | UGAGGUAGUAAGUUGUAUUGUU | ||||
| hsa-let-7gSZ | UGAGGUAGUAGUUUGUACAGU | ||||
| hsa-let-7ibrain | UGAGGUAGUAGUUUGUGCUGU | ||||
| miR-101 | 118 | 124 | 1A | hsa-miR-101 | UACAGUACUGUGAUAACUGAAG |
| miR-124.1 | 2541 | 2547 | m8 | hsa-miR-124a | UUAAGGCACGCGGUGAAUGCCA |
| miR-124/506 | 2541 | 2547 | 1A | hsa-miR-506 | UAAGGCACCCUUCUGAGUAGA |
| hsa-miR-124brain | UAAGGCACGCGGUGAAUGCC | ||||
| miR-129-5p | 196 | 202 | 1A | hsa-miR-129brain | CUUUUUGCGGUCUGGGCUUGC |
| hsa-miR-129-5p | CUUUUUGCGGUCUGGGCUUGCU | ||||
| miR-139 | 119 | 126 | 1A,m8 | hsa-miR-139brain | UCUACAGUGCACGUGUCU |
| miR-142-5p | 254 | 260 | 1A | hsa-miR-142-5p | CAUAAAGUAGAAAGCACUAC |
| hsa-miR-142-5p | CAUAAAGUAGAAAGCACUAC | ||||
| hsa-miR-142-5p | CAUAAAGUAGAAAGCACUAC | ||||
| miR-144 | 117 | 124 | 1A,m8 | hsa-miR-144 | UACAGUAUAGAUGAUGUACUAG |
| miR-153 | 183 | 190 | 1A,m8 | hsa-miR-153 | UUGCAUAGUCACAAAAGUGA |
| miR-186 | 2326 | 2332 | 1A | hsa-miR-186 | CAAAGAAUUCUCCUUUUGGGCUU |
| hsa-miR-186 | CAAAGAAUUCUCCUUUUGGGCUU | ||||
| miR-190 | 2517 | 2524 | 1A,m8 | hsa-miR-190 | UGAUAUGUUUGAUAUAUUAGGU |
| miR-194 | 1711 | 1717 | m8 | hsa-miR-194 | UGUAACAGCAACUCCAUGUGGA |
| miR-200bc/429 | 2342 | 2348 | 1A | hsa-miR-200b | UAAUACUGCCUGGUAAUGAUGAC |
| hsa-miR-200c | UAAUACUGCCGGGUAAUGAUGG | ||||
| hsa-miR-429 | UAAUACUGUCUGGUAAAACCGU | ||||
| miR-30-5p | 72 | 78 | 1A | hsa-miR-30a-5p | UGUAAACAUCCUCGACUGGAAG |
| hsa-miR-30cbrain | UGUAAACAUCCUACACUCUCAGC | ||||
| hsa-miR-30dSZ | UGUAAACAUCCCCGACUGGAAG | ||||
| hsa-miR-30bSZ | UGUAAACAUCCUACACUCAGCU | ||||
| hsa-miR-30e-5p | UGUAAACAUCCUUGACUGGA | ||||
| miR-31 | 244 | 250 | 1A | hsa-miR-31 | AGGCAAGAUGCUGGCAUAGCUG |
| miR-320 | 2272 | 2278 | 1A | hsa-miR-320 | AAAAGCUGGGUUGAGAGGGCGAA |
| hsa-miR-320 | AAAAGCUGGGUUGAGAGGGCGAA | ||||
| miR-369-3p | 2553 | 2560 | 1A,m8 | hsa-miR-369-3p | AAUAAUACAUGGUUGAUCUUU |
| miR-374 | 2554 | 2560 | m8 | hsa-miR-374 | UUAUAAUACAACCUGAUAAGUG |
| miR-421 | 413 | 419 | 1A | hsa-miR-421 | GGCCUCAUUAAAUGUUUGUUG |
| miR-448 | 184 | 190 | 1A | hsa-miR-448 | UUGCAUAUGUAGGAUGUCCCAU |
| miR-493-5p | 192 | 198 | 1A | hsa-miR-493-5p | UUGUACAUGGUAGGCUUUCAUU |
| miR-494 | 819 | 825 | m8 | hsa-miR-494brain | UGAAACAUACACGGGAAACCUCUU |
| miR-495 | 155 | 161 | 1A | hsa-miR-495brain | AAACAAACAUGGUGCACUUCUUU |
| miR-496 | 218 | 225 | 1A,m8 | hsa-miR-496 | AUUACAUGGCCAAUCUC |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.