Gene Page: DNMT3B
Summary ?
| GeneID | 1789 |
| Symbol | DNMT3B |
| Synonyms | ICF|ICF1|M.HsaIIIB |
| Description | DNA (cytosine-5-)-methyltransferase 3 beta |
| Reference | MIM:602900|HGNC:HGNC:2979|Ensembl:ENSG00000088305|HPRD:04209|Vega:OTTHUMG00000032226 |
| Gene type | protein-coding |
| Map location | 20q11.2 |
| Pascal p-value | 0.094 |
| Fetal beta | -0.24 |
| eGene | Cerebellar Hemisphere Cerebellum |
| Support | Chromatin Remodeling Genes |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenics,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
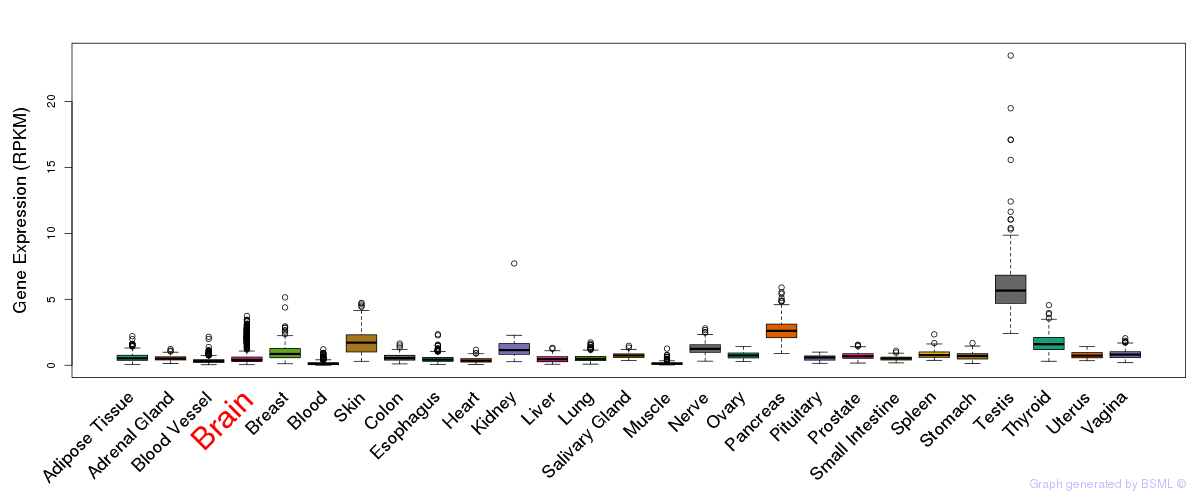
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| LRRC41 | 0.93 | 0.93 |
| KLHL22 | 0.92 | 0.92 |
| EDC4 | 0.91 | 0.91 |
| USP19 | 0.91 | 0.90 |
| USP21 | 0.91 | 0.92 |
| ZDHHC18 | 0.91 | 0.91 |
| PSMD3 | 0.91 | 0.91 |
| ZNF574 | 0.91 | 0.92 |
| DTX3 | 0.91 | 0.92 |
| TSC2 | 0.91 | 0.91 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.31 | -0.79 | -0.83 |
| MT-CO2 | -0.78 | -0.83 |
| AF347015.27 | -0.77 | -0.84 |
| AF347015.8 | -0.77 | -0.84 |
| MT-CYB | -0.76 | -0.82 |
| AF347015.21 | -0.76 | -0.89 |
| AF347015.33 | -0.74 | -0.80 |
| AF347015.15 | -0.73 | -0.80 |
| AF347015.2 | -0.72 | -0.79 |
| AF347015.26 | -0.70 | -0.78 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| CBX1 | CBX | HP1-BETA | HP1Hs-beta | HP1Hsbeta | M31 | MOD1 | p25beta | chromobox homolog 1 (HP1 beta homolog Drosophila ) | DNMT3B interacts with HP1-beta. This interaction was modeled on a demonstrated interaction between mouse Dnmt3b1 and human HP1-beta. | BIND | 15120635 |
| CBX1 | CBX | HP1-BETA | HP1Hs-beta | HP1Hsbeta | M31 | MOD1 | p25beta | chromobox homolog 1 (HP1 beta homolog Drosophila ) | - | HPRD,BioGRID | 12867029 |
| CBX3 | HECH | HP1-GAMMA | HP1Hs-gamma | chromobox homolog 3 (HP1 gamma homolog, Drosophila) | DNMT3B interacts with HP1-gamma. This interaction was modeled on a demonstrated interaction between mouse Dnmt3b1 and human HP1-gamma. | BIND | 15120635 |
| CBX5 | HP1 | HP1A | chromobox homolog 5 (HP1 alpha homolog, Drosophila) | - | HPRD,BioGRID | 12867029 |
| CBX5 | HP1 | HP1A | chromobox homolog 5 (HP1 alpha homolog, Drosophila) | DNMT3B interacts with HP1-alpha. This interaction was modeled on a demonstrated interaction between mouse Dnmt3b1 and human HP1-alpha. | BIND | 15120635 |
| DNMT1 | AIM | CXXC9 | DNMT | FLJ16293 | MCMT | MGC104992 | DNA (cytosine-5-)-methyltransferase 1 | DNMT3B interacts with DNMT1. | BIND | 15120635 |
| DNMT1 | AIM | CXXC9 | DNMT | FLJ16293 | MCMT | MGC104992 | DNA (cytosine-5-)-methyltransferase 1 | - | HPRD,BioGRID | 12145218 |
| DNMT1 | AIM | CXXC9 | DNMT | FLJ16293 | MCMT | MGC104992 | DNA (cytosine-5-)-methyltransferase 1 | hDNMT3b interacts with hDNMT1. | BIND | 12145218 |
| DNMT3A | DNMT3A2 | M.HsaIIIA | DNA (cytosine-5-)-methyltransferase 3 alpha | - | HPRD,BioGRID | 12145218 |
| DNMT3L | MGC1090 | DNA (cytosine-5-)-methyltransferase 3-like | - | HPRD,BioGRID | 11934864 |
| DNMT3L | MGC1090 | DNA (cytosine-5-)-methyltransferase 3-like | DNMT3L interacts with Dnmt3b. This interaction was modeled on a demonstrated interaction between human DNMT3L and mouse Dnmt3a. | BIND | 15105426 |
| HDAC1 | DKFZp686H12203 | GON-10 | HD1 | RPD3 | RPD3L1 | histone deacetylase 1 | Co-purification | BioGRID | 15148359 |
| HDAC1 | DKFZp686H12203 | GON-10 | HD1 | RPD3 | RPD3L1 | histone deacetylase 1 | - | HPRD | 12202768 |
| HDAC1 | DKFZp686H12203 | GON-10 | HD1 | RPD3 | RPD3L1 | histone deacetylase 1 | DNMT3B interacts with HDAC1. | BIND | 15120635 |
| HDAC2 | RPD3 | YAF1 | histone deacetylase 2 | DNMT3B interacts with HDAC2. | BIND | 15120635 |
| KIF4A | FLJ12530 | FLJ12655 | FLJ14204 | FLJ20631 | HSA271784 | KIF4 | KIF4-G1 | kinesin family member 4A | Co-purification Reconstituted Complex | BioGRID | 15148359 |
| NCAPG | CAPG | CHCG | FLJ12450 | HCAP-G | MGC126525 | NY-MEL-3 | non-SMC condensin I complex, subunit G | - | HPRD,BioGRID | 15148359 |
| RARB | HAP | NR1B2 | RRB2 | retinoic acid receptor, beta | DNMT3B interacts with RARB (RAR-beta-2) gene promoter sequences. | BIND | 15688037 |
| SIN3A | DKFZp434K2235 | FLJ90319 | KIAA0700 | SIN3 homolog A, transcription regulator (yeast) | Co-purification | BioGRID | 15148359 |
| SMARCA5 | ISWI | SNF2H | WCRF135 | hISWI | hSNF2H | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 5 | Co-purification | BioGRID | 15148359 |
| SMARCA5 | ISWI | SNF2H | WCRF135 | hISWI | hSNF2H | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 5 | DNMT3B interacts with hSNF2H. | BIND | 15120635 |
| SMC2 | CAP-E | CAPE | FLJ10093 | SMC2L1 | hCAP-E | structural maintenance of chromosomes 2 | Co-purification Reconstituted Complex | BioGRID | 15148359 |
| SMC4 | CAPC | SMC4L1 | hCAP-C | structural maintenance of chromosomes 4 | Co-purification | BioGRID | 15148359 |
| SUMO1 | DAP-1 | GMP1 | OFC10 | PIC1 | SENP2 | SMT3 | SMT3C | SMT3H3 | SUMO-1 | UBL1 | SMT3 suppressor of mif two 3 homolog 1 (S. cerevisiae) | Dnmt3b interacts with SUMO-1. | BIND | 11735126 |
| SUMO1 | DAP-1 | GMP1 | OFC10 | PIC1 | SENP2 | SMT3 | SMT3C | SMT3H3 | SUMO-1 | UBL1 | SMT3 suppressor of mif two 3 homolog 1 (S. cerevisiae) | - | HPRD,BioGRID | 11735126 |
| UBE2I | C358B7.1 | P18 | UBC9 | ubiquitin-conjugating enzyme E2I (UBC9 homolog, yeast) | Dnmt3b interacts with Ubc9. | BIND | 11735126 |
| UBE2I | C358B7.1 | P18 | UBC9 | ubiquitin-conjugating enzyme E2I (UBC9 homolog, yeast) | - | HPRD,BioGRID | 11735126 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG CYSTEINE AND METHIONINE METABOLISM | 34 | 24 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP | 1382 | 904 | All SZGR 2.0 genes in this pathway |
| TIEN INTESTINE PROBIOTICS 24HR UP | 557 | 331 | All SZGR 2.0 genes in this pathway |
| JAATINEN HEMATOPOIETIC STEM CELL UP | 316 | 190 | All SZGR 2.0 genes in this pathway |
| LIU CDX2 TARGETS UP | 36 | 22 | All SZGR 2.0 genes in this pathway |
| PEREZ TP53 TARGETS | 1174 | 695 | All SZGR 2.0 genes in this pathway |
| LINDGREN BLADDER CANCER HIGH RECURRENCE | 49 | 31 | All SZGR 2.0 genes in this pathway |
| ROSTY CERVICAL CANCER PROLIFERATION CLUSTER | 140 | 73 | All SZGR 2.0 genes in this pathway |
| KORKOLA TERATOMA | 39 | 25 | All SZGR 2.0 genes in this pathway |
| NUYTTEN NIPP1 TARGETS UP | 769 | 437 | All SZGR 2.0 genes in this pathway |
| NUYTTEN EZH2 TARGETS UP | 1037 | 673 | All SZGR 2.0 genes in this pathway |
| BUYTAERT PHOTODYNAMIC THERAPY STRESS UP | 811 | 508 | All SZGR 2.0 genes in this pathway |
| BENPORATH ES 1 | 379 | 235 | All SZGR 2.0 genes in this pathway |
| ZHANG RESPONSE TO IKK INHIBITOR AND TNF DN | 103 | 64 | All SZGR 2.0 genes in this pathway |
| ROSS ACUTE MYELOID LEUKEMIA CBF | 82 | 57 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS STEM CELL AND PROGENITOR | 681 | 420 | All SZGR 2.0 genes in this pathway |
| SANSOM APC TARGETS REQUIRE MYC | 210 | 123 | All SZGR 2.0 genes in this pathway |
| AMUNDSON GAMMA RADIATION RESPONSE | 40 | 25 | All SZGR 2.0 genes in this pathway |
| CONRAD STEM CELL | 39 | 27 | All SZGR 2.0 genes in this pathway |
| MUELLER PLURINET | 299 | 189 | All SZGR 2.0 genes in this pathway |
| KORKOLA EMBRYONIC CARCINOMA VS SEMINOMA UP | 22 | 15 | All SZGR 2.0 genes in this pathway |
| WONG ADULT TISSUE STEM MODULE | 721 | 492 | All SZGR 2.0 genes in this pathway |
| ZEMBUTSU SENSITIVITY TO DOXORUBICIN | 17 | 10 | All SZGR 2.0 genes in this pathway |
| WIEDERSCHAIN TARGETS OF BMI1 AND PCGF2 | 57 | 39 | All SZGR 2.0 genes in this pathway |
| TORCHIA TARGETS OF EWSR1 FLI1 FUSION DN | 321 | 200 | All SZGR 2.0 genes in this pathway |
| GOBERT OLIGODENDROCYTE DIFFERENTIATION DN | 1080 | 713 | All SZGR 2.0 genes in this pathway |
| ZWANG DOWN BY 2ND EGF PULSE | 293 | 119 | All SZGR 2.0 genes in this pathway |