Gene Page: DPYS
Summary ?
| GeneID | 1807 |
| Symbol | DPYS |
| Synonyms | DHP|DHPase |
| Description | dihydropyrimidinase |
| Reference | MIM:613326|HGNC:HGNC:3013|Ensembl:ENSG00000147647|HPRD:01960|Vega:OTTHUMG00000164891 |
| Gene type | protein-coding |
| Map location | 8q22 |
| Pascal p-value | 0.96 |
| Fetal beta | -2.651 |
| DMG | 1 (# studies) |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg17087640 | 8 | 105479215 | DPYS | 2.57E-5 | 0.349 | 0.017 | DMG:Wockner_2014 |
Section II. Transcriptome annotation
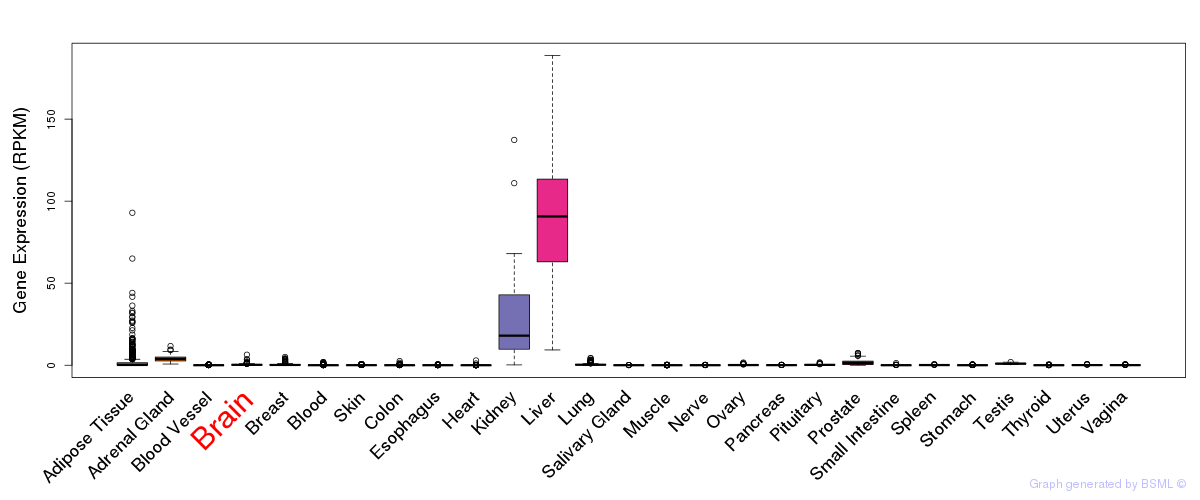
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| NIT1 | 0.88 | 0.84 |
| MPI | 0.85 | 0.83 |
| PDCD6 | 0.85 | 0.83 |
| POLR2C | 0.84 | 0.77 |
| APEH | 0.84 | 0.79 |
| CAPZB | 0.83 | 0.81 |
| WDR45 | 0.81 | 0.78 |
| VRK3 | 0.81 | 0.76 |
| PHKG2 | 0.81 | 0.78 |
| ARFIP2 | 0.81 | 0.76 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.21 | -0.59 | -0.41 |
| AF347015.18 | -0.58 | -0.44 |
| MT-ATP8 | -0.54 | -0.40 |
| AF347015.8 | -0.54 | -0.36 |
| AF347015.2 | -0.54 | -0.35 |
| MT-CO2 | -0.52 | -0.34 |
| MT-CYB | -0.51 | -0.35 |
| AF347015.26 | -0.51 | -0.35 |
| AF347015.31 | -0.50 | -0.35 |
| AF347015.27 | -0.49 | -0.39 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG PYRIMIDINE METABOLISM | 98 | 53 | All SZGR 2.0 genes in this pathway |
| KEGG BETA ALANINE METABOLISM | 22 | 16 | All SZGR 2.0 genes in this pathway |
| KEGG PANTOTHENATE AND COA BIOSYNTHESIS | 16 | 9 | All SZGR 2.0 genes in this pathway |
| KEGG DRUG METABOLISM OTHER ENZYMES | 51 | 27 | All SZGR 2.0 genes in this pathway |
| REACTOME PYRIMIDINE CATABOLISM | 12 | 5 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF NUCLEOTIDES | 72 | 45 | All SZGR 2.0 genes in this pathway |
| REACTOME PYRIMIDINE METABOLISM | 24 | 13 | All SZGR 2.0 genes in this pathway |
| SHETH LIVER CANCER VS TXNIP LOSS PAM4 | 261 | 153 | All SZGR 2.0 genes in this pathway |
| LEE LIVER CANCER DENA DN | 74 | 45 | All SZGR 2.0 genes in this pathway |
| FLECHNER BIOPSY KIDNEY TRANSPLANT REJECTED VS OK DN | 546 | 351 | All SZGR 2.0 genes in this pathway |
| HSIAO LIVER SPECIFIC GENES | 244 | 154 | All SZGR 2.0 genes in this pathway |
| YAMAZAKI TCEB3 TARGETS DN | 215 | 132 | All SZGR 2.0 genes in this pathway |
| IWANAGA CARCINOGENESIS BY KRAS PTEN UP | 181 | 112 | All SZGR 2.0 genes in this pathway |
| RIZKI TUMOR INVASIVENESS 3D DN | 270 | 181 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER CANCER DN | 540 | 340 | All SZGR 2.0 genes in this pathway |
| LEE LIVER CANCER SURVIVAL UP | 185 | 112 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS UP | 1305 | 895 | All SZGR 2.0 genes in this pathway |
| CHIANG LIVER CANCER SUBCLASS UNANNOTATED UP | 85 | 50 | All SZGR 2.0 genes in this pathway |
| CAIRO LIVER DEVELOPMENT DN | 222 | 141 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN MEF HCP WITH H3K27ME3 | 590 | 403 | All SZGR 2.0 genes in this pathway |