Gene Page: DR1
Summary ?
| GeneID | 1810 |
| Symbol | DR1 |
| Synonyms | NC2|NC2-BETA|NC2B |
| Description | down-regulator of transcription 1 |
| Reference | MIM:601482|HGNC:HGNC:3017|HPRD:03284| |
| Gene type | protein-coding |
| Map location | 1p22.1 |
| Pascal p-value | 0.683 |
| Fetal beta | 0.626 |
| eGene | Caudate basal ganglia Frontal Cortex BA9 |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Expression | Meta-analysis of gene expression | P value: 1.996 | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenics,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
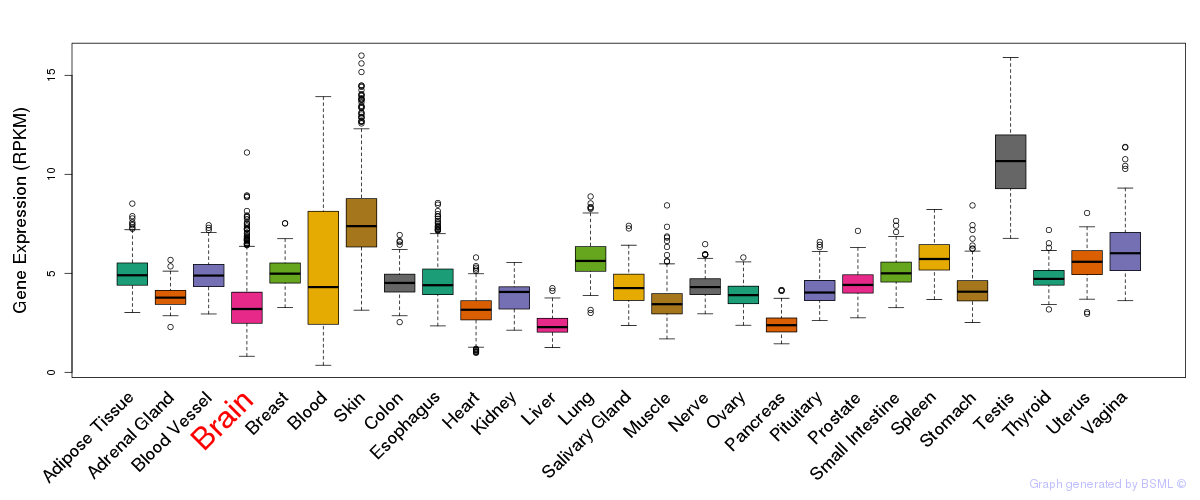
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| ATXN2L | 0.94 | 0.95 |
| PTPN23 | 0.93 | 0.95 |
| CIC | 0.93 | 0.95 |
| ZMIZ2 | 0.93 | 0.93 |
| PCNXL3 | 0.93 | 0.93 |
| BAT2 | 0.93 | 0.95 |
| MGRN1 | 0.92 | 0.93 |
| TSC2 | 0.91 | 0.92 |
| ZNF142 | 0.91 | 0.92 |
| ZC3H3 | 0.91 | 0.93 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.31 | -0.71 | -0.77 |
| MT-CO2 | -0.70 | -0.77 |
| AF347015.21 | -0.69 | -0.84 |
| GNG11 | -0.68 | -0.79 |
| S100B | -0.67 | -0.74 |
| C5orf53 | -0.67 | -0.69 |
| C1orf54 | -0.67 | -0.83 |
| HIGD1B | -0.66 | -0.75 |
| AC021016.1 | -0.66 | -0.74 |
| MT-CYB | -0.65 | -0.71 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0003714 | transcription corepressor activity | TAS | 8670811 | |
| GO:0008134 | transcription factor binding | TAS | 1339312 | |
| GO:0043565 | sequence-specific DNA binding | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0000122 | negative regulation of transcription from RNA polymerase II promoter | TAS | 8670811 | |
| GO:0006355 | regulation of transcription, DNA-dependent | IEA | - | |
| GO:0006350 | transcription | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005622 | intracellular | IEA | - | |
| GO:0005634 | nucleus | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP | 1382 | 904 | All SZGR 2.0 genes in this pathway |
| BASAKI YBX1 TARGETS UP | 290 | 177 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA1 PCC NETWORK | 1652 | 1023 | All SZGR 2.0 genes in this pathway |
| PUJANA ATM PCC NETWORK | 1442 | 892 | All SZGR 2.0 genes in this pathway |
| WEI MYCN TARGETS WITH E BOX | 795 | 478 | All SZGR 2.0 genes in this pathway |
| BENPORATH CYCLING GENES | 648 | 385 | All SZGR 2.0 genes in this pathway |
| BENPORATH ES CORE NINE CORRELATED | 100 | 68 | All SZGR 2.0 genes in this pathway |
| SMITH TERT TARGETS UP | 145 | 91 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 16HR UP | 225 | 139 | All SZGR 2.0 genes in this pathway |
| MA PITUITARY FETAL VS ADULT DN | 19 | 17 | All SZGR 2.0 genes in this pathway |
| GENTILE UV HIGH DOSE DN | 312 | 203 | All SZGR 2.0 genes in this pathway |
| SESTO RESPONSE TO UV C0 | 107 | 72 | All SZGR 2.0 genes in this pathway |
| WELCSH BRCA1 TARGETS UP | 198 | 132 | All SZGR 2.0 genes in this pathway |
| GENTILE RESPONSE CLUSTER D3 | 61 | 39 | All SZGR 2.0 genes in this pathway |
| WINTER HYPOXIA METAGENE | 242 | 168 | All SZGR 2.0 genes in this pathway |
| SHEDDEN LUNG CANCER POOR SURVIVAL A6 | 456 | 285 | All SZGR 2.0 genes in this pathway |
| BOYAULT LIVER CANCER SUBCLASS G3 UP | 188 | 121 | All SZGR 2.0 genes in this pathway |
| PYEON CANCER HEAD AND NECK VS CERVICAL UP | 193 | 95 | All SZGR 2.0 genes in this pathway |
| WHITFIELD CELL CYCLE G2 M | 216 | 124 | All SZGR 2.0 genes in this pathway |
| JOHNSTONE PARVB TARGETS 3 DN | 918 | 550 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-101 | 1724 | 1731 | 1A,m8 | hsa-miR-101 | UACAGUACUGUGAUAACUGAAG |
| miR-122 | 747 | 753 | 1A | hsa-miR-122a | UGGAGUGUGACAAUGGUGUUUGU |
| miR-137 | 1924 | 1931 | 1A,m8 | hsa-miR-137 | UAUUGCUUAAGAAUACGCGUAG |
| miR-141/200a | 282 | 289 | 1A,m8 | hsa-miR-141 | UAACACUGUCUGGUAAAGAUGG |
| hsa-miR-200a | UAACACUGUCUGGUAACGAUGU | ||||
| miR-144 | 1725 | 1731 | 1A | hsa-miR-144 | UACAGUAUAGAUGAUGUACUAG |
| miR-182 | 1366 | 1373 | 1A,m8 | hsa-miR-182 | UUUGGCAAUGGUAGAACUCACA |
| miR-194 | 285 | 291 | 1A | hsa-miR-194 | UGUAACAGCAACUCCAUGUGGA |
| miR-203.1 | 697 | 704 | 1A,m8 | hsa-miR-203 | UGAAAUGUUUAGGACCACUAG |
| miR-224 | 1876 | 1882 | m8 | hsa-miR-224 | CAAGUCACUAGUGGUUCCGUUUA |
| miR-320 | 1067 | 1073 | 1A | hsa-miR-320 | AAAAGCUGGGUUGAGAGGGCGAA |
| hsa-miR-320 | AAAAGCUGGGUUGAGAGGGCGAA | ||||
| miR-369-3p | 1855 | 1862 | 1A,m8 | hsa-miR-369-3p | AAUAAUACAUGGUUGAUCUUU |
| miR-374 | 1856 | 1862 | m8 | hsa-miR-374 | UUAUAAUACAACCUGAUAAGUG |
| miR-409-3p | 557 | 563 | 1A | hsa-miR-409-3p | CGAAUGUUGCUCGGUGAACCCCU |
| miR-490 | 1826 | 1832 | 1A | hsa-miR-490 | CAACCUGGAGGACUCCAUGCUG |
| miR-496 | 161 | 168 | 1A,m8 | hsa-miR-496 | AUUACAUGGCCAAUCUC |
| miR-9 | 1668 | 1674 | m8 | hsa-miR-9SZ | UCUUUGGUUAUCUAGCUGUAUGA |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.