Gene Page: DRD5
Summary ?
| GeneID | 1816 |
| Symbol | DRD5 |
| Synonyms | DBDR|DRD1B|DRD1L2 |
| Description | dopamine receptor D5 |
| Reference | MIM:126453|HGNC:HGNC:3026|Ensembl:ENSG00000169676|HPRD:00542|Vega:OTTHUMG00000128489 |
| Gene type | protein-coding |
| Map location | 4p16.1 |
| Pascal p-value | 0.039 |
| Sherlock p-value | 0.761 |
| Fetal beta | -2.75 |
| eGene | Myers' cis & trans |
| Support | DOPAMINE |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| ADT:Sun_2012 | Systematic Investigation of Antipsychotic Drugs and Their Targets | A total of 382 drug-target associations involving 43 antipsychotic drugs and 49 target genes. | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Association | A combined odds ratio method (Sun et al. 2008), association studies | 2 | Link to SZGene |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenics,schizotypy,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 5 |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs7142486 | chr14 | 84504067 | DRD5 | 1816 | 0.09 | trans |
Section II. Transcriptome annotation
General gene expression (GTEx)
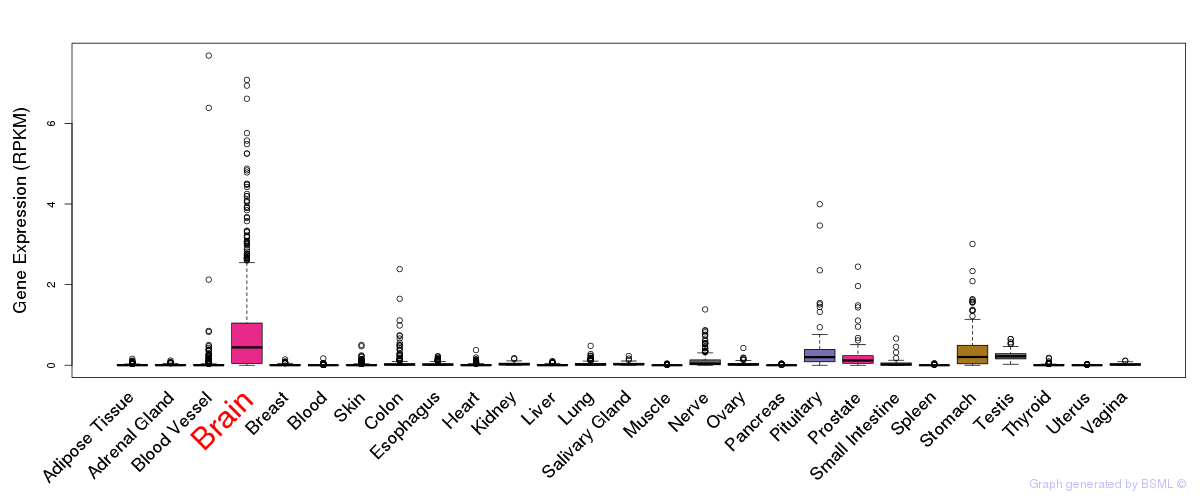
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AC110491.1 | 0.60 | 0.47 |
| CRYBG3 | 0.60 | 0.51 |
| CFTR | 0.58 | 0.45 |
| DLC1 | 0.57 | 0.46 |
| RHOQ | 0.56 | 0.44 |
| ANO3 | 0.55 | 0.45 |
| SLC1A2 | 0.55 | 0.39 |
| TJP1 | 0.54 | 0.52 |
| SCN4B | 0.54 | 0.42 |
| ITGAV | 0.54 | 0.43 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| RPL35 | -0.31 | -0.41 |
| RPL36 | -0.30 | -0.41 |
| RPS7 | -0.29 | -0.34 |
| PFDN5 | -0.29 | -0.34 |
| BCL7C | -0.29 | -0.37 |
| RPLP1 | -0.28 | -0.37 |
| RPS14 | -0.28 | -0.34 |
| FAU | -0.28 | -0.36 |
| RPL31 | -0.28 | -0.35 |
| SCNM1 | -0.28 | -0.30 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0001588 | dopamine receptor coupled via Gs | TAS | Neurotransmitter, dopamine (GO term level: 9) | 9457173 |
| GO:0004872 | receptor activity | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0060158 | dopamine receptor, phospholipase C activating pathway | TAS | dopamine (GO term level: 10) | 9457173 |
| GO:0007268 | synaptic transmission | NAS | neuron, Synap, Neurotransmitter (GO term level: 6) | 7727453 |
| GO:0007191 | dopamine receptor, adenylate cyclase activating pathway | IDA | dopamine (GO term level: 12) | 1826762 |
| GO:0001963 | synaptic transmission, dopaminergic | NAS | neuron, Synap, Neurotransmitter, dopamine (GO term level: 8) | 11036203 |
| GO:0007190 | activation of adenylate cyclase activity | TAS | 9457173 | |
| GO:0007186 | G-protein coupled receptor protein signaling pathway | IEA | - | |
| GO:0007165 | signal transduction | IEA | - | |
| GO:0006800 | oxygen and reactive oxygen species metabolic process | IDA | 16352863 | |
| GO:0006874 | cellular calcium ion homeostasis | TAS | 9457173 | |
| GO:0033861 | negative regulation of NAD(P)H oxidase activity | IDA | 16352863 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005886 | plasma membrane | IDA | 10531415 |11500503 | |
| GO:0005887 | integral to plasma membrane | NAS | 1826762 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG CALCIUM SIGNALING PATHWAY | 178 | 134 | All SZGR 2.0 genes in this pathway |
| KEGG NEUROACTIVE LIGAND RECEPTOR INTERACTION | 272 | 195 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY GPCR | 920 | 449 | All SZGR 2.0 genes in this pathway |
| REACTOME CLASS A1 RHODOPSIN LIKE RECEPTORS | 305 | 177 | All SZGR 2.0 genes in this pathway |
| REACTOME AMINE LIGAND BINDING RECEPTORS | 38 | 33 | All SZGR 2.0 genes in this pathway |
| REACTOME GPCR DOWNSTREAM SIGNALING | 805 | 368 | All SZGR 2.0 genes in this pathway |
| REACTOME G ALPHA S SIGNALLING EVENTS | 121 | 82 | All SZGR 2.0 genes in this pathway |
| REACTOME GPCR LIGAND BINDING | 408 | 246 | All SZGR 2.0 genes in this pathway |
| DARWICHE SKIN TUMOR PROMOTER UP | 142 | 96 | All SZGR 2.0 genes in this pathway |
| DARWICHE PAPILLOMA RISK LOW UP | 162 | 104 | All SZGR 2.0 genes in this pathway |
| DARWICHE PAPILLOMA RISK HIGH UP | 147 | 101 | All SZGR 2.0 genes in this pathway |
| DARWICHE SQUAMOUS CELL CARCINOMA UP | 146 | 104 | All SZGR 2.0 genes in this pathway |
| BENPORATH SUZ12 TARGETS | 1038 | 678 | All SZGR 2.0 genes in this pathway |
| BENPORATH EED TARGETS | 1062 | 725 | All SZGR 2.0 genes in this pathway |
| BENPORATH ES WITH H3K27ME3 | 1118 | 744 | All SZGR 2.0 genes in this pathway |
| BENPORATH PRC2 TARGETS | 652 | 441 | All SZGR 2.0 genes in this pathway |
| MOREAUX MULTIPLE MYELOMA BY TACI UP | 412 | 249 | All SZGR 2.0 genes in this pathway |
| XU GH1 EXOGENOUS TARGETS UP | 85 | 50 | All SZGR 2.0 genes in this pathway |
| XU GH1 AUTOCRINE TARGETS UP | 268 | 157 | All SZGR 2.0 genes in this pathway |
| MEISSNER NPC HCP WITH H3 UNMETHYLATED | 536 | 296 | All SZGR 2.0 genes in this pathway |
| MEISSNER BRAIN HCP WITH H3K4ME3 AND H3K27ME3 | 1069 | 729 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN MCV6 HCP WITH H3K27ME3 | 435 | 318 | All SZGR 2.0 genes in this pathway |
| MARTENS TRETINOIN RESPONSE UP | 857 | 456 | All SZGR 2.0 genes in this pathway |
| ZWANG DOWN BY 2ND EGF PULSE | 293 | 119 | All SZGR 2.0 genes in this pathway |