Gene Page: DUSP3
Summary ?
| GeneID | 1845 |
| Symbol | DUSP3 |
| Synonyms | VHR |
| Description | dual specificity phosphatase 3 |
| Reference | MIM:600183|HGNC:HGNC:3069|Ensembl:ENSG00000108861|HPRD:02553|Vega:OTTHUMG00000180889 |
| Gene type | protein-coding |
| Map location | 17q21 |
| Pascal p-value | 0.442 |
| Sherlock p-value | 6.393E-4 |
| Fetal beta | -1.705 |
| eGene | Myers' cis & trans |
| Support | G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-FULL G2Cdb.humanPSD G2Cdb.humanPSP |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.0357 |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs4450223 | chr12 | 46418660 | DUSP3 | 1845 | 0.06 | trans |
Section II. Transcriptome annotation
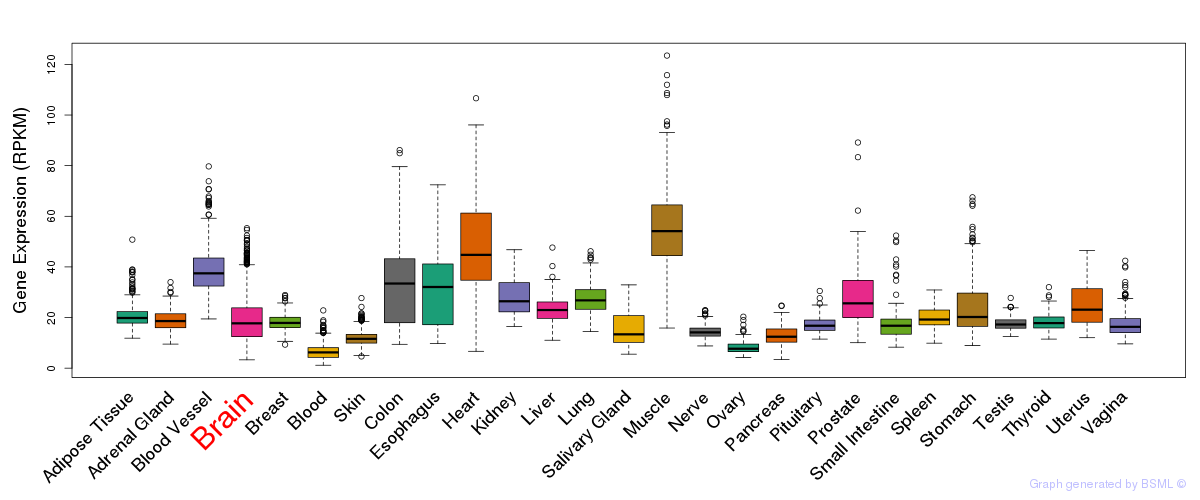
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| MTA1 | 0.97 | 0.97 |
| EHMT2 | 0.97 | 0.97 |
| RBM10 | 0.97 | 0.97 |
| U2AF2 | 0.97 | 0.97 |
| DCAF15 | 0.96 | 0.96 |
| CXXC1 | 0.96 | 0.95 |
| WHSC2 | 0.96 | 0.96 |
| DAXX | 0.96 | 0.94 |
| ZNF777 | 0.95 | 0.96 |
| XAB2 | 0.95 | 0.95 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.31 | -0.78 | -0.90 |
| AF347015.27 | -0.77 | -0.89 |
| MT-CO2 | -0.77 | -0.90 |
| C5orf53 | -0.76 | -0.79 |
| AF347015.33 | -0.75 | -0.87 |
| MT-CYB | -0.74 | -0.87 |
| S100B | -0.73 | -0.83 |
| AF347015.8 | -0.73 | -0.88 |
| FXYD1 | -0.71 | -0.82 |
| COPZ2 | -0.71 | -0.79 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0004725 | protein tyrosine phosphatase activity | EXP | 17322878 | |
| GO:0004725 | protein tyrosine phosphatase activity | IEA | - | |
| GO:0004725 | protein tyrosine phosphatase activity | TAS | 1281549 | |
| GO:0016787 | hydrolase activity | IEA | - | |
| GO:0016791 | phosphatase activity | IEA | - | |
| GO:0008138 | protein tyrosine/serine/threonine phosphatase activity | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006470 | protein amino acid dephosphorylation | IEA | - | |
| GO:0006470 | protein amino acid dephosphorylation | TAS | 1281549 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005654 | nucleoplasm | EXP | 17322878 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG MAPK SIGNALING PATHWAY | 267 | 205 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALLING BY NGF | 217 | 167 | All SZGR 2.0 genes in this pathway |
| REACTOME TRIF MEDIATED TLR3 SIGNALING | 74 | 54 | All SZGR 2.0 genes in this pathway |
| REACTOME NGF SIGNALLING VIA TRKA FROM THE PLASMA MEMBRANE | 137 | 105 | All SZGR 2.0 genes in this pathway |
| REACTOME NUCLEAR EVENTS KINASE AND TRANSCRIPTION FACTOR ACTIVATION | 24 | 20 | All SZGR 2.0 genes in this pathway |
| REACTOME ERKS ARE INACTIVATED | 12 | 9 | All SZGR 2.0 genes in this pathway |
| REACTOME ERK MAPK TARGETS | 21 | 17 | All SZGR 2.0 genes in this pathway |
| REACTOME MAP KINASE ACTIVATION IN TLR CASCADE | 50 | 37 | All SZGR 2.0 genes in this pathway |
| REACTOME MAPK TARGETS NUCLEAR EVENTS MEDIATED BY MAP KINASES | 30 | 24 | All SZGR 2.0 genes in this pathway |
| REACTOME TRAF6 MEDIATED INDUCTION OF NFKB AND MAP KINASES UPON TLR7 8 OR 9 ACTIVATION | 77 | 57 | All SZGR 2.0 genes in this pathway |
| REACTOME NFKB AND MAP KINASES ACTIVATION MEDIATED BY TLR4 SIGNALING REPERTOIRE | 72 | 53 | All SZGR 2.0 genes in this pathway |
| REACTOME MYD88 MAL CASCADE INITIATED ON PLASMA MEMBRANE | 83 | 63 | All SZGR 2.0 genes in this pathway |
| REACTOME INNATE IMMUNE SYSTEM | 279 | 178 | All SZGR 2.0 genes in this pathway |
| REACTOME ACTIVATED TLR4 SIGNALLING | 93 | 69 | All SZGR 2.0 genes in this pathway |
| REACTOME IMMUNE SYSTEM | 933 | 616 | All SZGR 2.0 genes in this pathway |
| REACTOME TOLL RECEPTOR CASCADES | 118 | 84 | All SZGR 2.0 genes in this pathway |
| WATANABE RECTAL CANCER RADIOTHERAPY RESPONSIVE UP | 108 | 67 | All SZGR 2.0 genes in this pathway |
| ONKEN UVEAL MELANOMA DN | 526 | 357 | All SZGR 2.0 genes in this pathway |
| WANG LMO4 TARGETS DN | 352 | 225 | All SZGR 2.0 genes in this pathway |
| WANG CLIM2 TARGETS DN | 186 | 114 | All SZGR 2.0 genes in this pathway |
| GARGALOVIC RESPONSE TO OXIDIZED PHOSPHOLIPIDS TURQUOISE UP | 79 | 50 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC1 TARGETS UP | 457 | 269 | All SZGR 2.0 genes in this pathway |
| CONCANNON APOPTOSIS BY EPOXOMICIN UP | 239 | 157 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 UP | 309 | 199 | All SZGR 2.0 genes in this pathway |
| GROSS HYPOXIA VIA HIF1A DN | 110 | 78 | All SZGR 2.0 genes in this pathway |
| NUYTTEN EZH2 TARGETS UP | 1037 | 673 | All SZGR 2.0 genes in this pathway |
| LOPEZ MBD TARGETS | 957 | 597 | All SZGR 2.0 genes in this pathway |
| RICKMAN TUMOR DIFFERENTIATED MODERATELY VS POORLY UP | 121 | 71 | All SZGR 2.0 genes in this pathway |
| BROCKE APOPTOSIS REVERSED BY IL6 | 144 | 98 | All SZGR 2.0 genes in this pathway |
| WONG IFNA2 RESISTANCE DN | 34 | 20 | All SZGR 2.0 genes in this pathway |
| FLECHNER BIOPSY KIDNEY TRANSPLANT REJECTED VS OK DN | 546 | 351 | All SZGR 2.0 genes in this pathway |
| FLECHNER BIOPSY KIDNEY TRANSPLANT OK VS DONOR UP | 555 | 346 | All SZGR 2.0 genes in this pathway |
| MOREAUX MULTIPLE MYELOMA BY TACI UP | 412 | 249 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 20HR UP | 240 | 152 | All SZGR 2.0 genes in this pathway |
| DEBIASI APOPTOSIS BY REOVIRUS INFECTION UP | 314 | 201 | All SZGR 2.0 genes in this pathway |
| LU AGING BRAIN DN | 153 | 120 | All SZGR 2.0 genes in this pathway |
| CUI TCF21 TARGETS 2 DN | 830 | 547 | All SZGR 2.0 genes in this pathway |
| DOUGLAS BMI1 TARGETS DN | 314 | 188 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 6HR DN | 911 | 527 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 24HR DN | 1011 | 592 | All SZGR 2.0 genes in this pathway |
| NAKAMURA METASTASIS | 47 | 28 | All SZGR 2.0 genes in this pathway |
| NAKAMURA METASTASIS MODEL UP | 45 | 26 | All SZGR 2.0 genes in this pathway |
| BREDEMEYER RAG SIGNALING NOT VIA ATM DN | 57 | 34 | All SZGR 2.0 genes in this pathway |
| BEIER GLIOMA STEM CELL DN | 66 | 42 | All SZGR 2.0 genes in this pathway |
| AGUIRRE PANCREATIC CANCER COPY NUMBER UP | 298 | 174 | All SZGR 2.0 genes in this pathway |
| ZHANG TLX TARGETS 36HR UP | 221 | 150 | All SZGR 2.0 genes in this pathway |
| ZHANG TLX TARGETS 60HR UP | 293 | 203 | All SZGR 2.0 genes in this pathway |
| ZHANG TLX TARGETS UP | 108 | 78 | All SZGR 2.0 genes in this pathway |
| BOYAULT LIVER CANCER SUBCLASS G3 UP | 188 | 121 | All SZGR 2.0 genes in this pathway |
| COULOUARN TEMPORAL TGFB1 SIGNATURE DN | 138 | 99 | All SZGR 2.0 genes in this pathway |
| SASSON RESPONSE TO GONADOTROPHINS UP | 91 | 59 | All SZGR 2.0 genes in this pathway |
| SASSON RESPONSE TO FORSKOLIN UP | 90 | 58 | All SZGR 2.0 genes in this pathway |
| KIM ALL DISORDERS CALB1 CORR UP | 548 | 370 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-124/506 | 3056 | 3062 | m8 | hsa-miR-506 | UAAGGCACCCUUCUGAGUAGA |
| hsa-miR-124brain | UAAGGCACGCGGUGAAUGCC | ||||
| miR-141/200a | 3434 | 3441 | 1A,m8 | hsa-miR-141 | UAACACUGUCUGGUAAAGAUGG |
| hsa-miR-200a | UAACACUGUCUGGUAACGAUGU | ||||
| miR-15/16/195/424/497 | 2110 | 2116 | m8 | hsa-miR-15abrain | UAGCAGCACAUAAUGGUUUGUG |
| hsa-miR-16brain | UAGCAGCACGUAAAUAUUGGCG | ||||
| hsa-miR-15bbrain | UAGCAGCACAUCAUGGUUUACA | ||||
| hsa-miR-195SZ | UAGCAGCACAGAAAUAUUGGC | ||||
| hsa-miR-424 | CAGCAGCAAUUCAUGUUUUGAA | ||||
| hsa-miR-497 | CAGCAGCACACUGUGGUUUGU | ||||
| miR-150 | 2476 | 2483 | 1A,m8 | hsa-miR-150 | UCUCCCAACCCUUGUACCAGUG |
| miR-153 | 428 | 435 | 1A,m8 | hsa-miR-153 | UUGCAUAGUCACAAAAGUGA |
| miR-370 | 2999 | 3005 | m8 | hsa-miR-370brain | GCCUGCUGGGGUGGAACCUGG |
| miR-384 | 74 | 80 | 1A | hsa-miR-384 | AUUCCUAGAAAUUGUUCAUA |
| miR-448 | 429 | 435 | 1A | hsa-miR-448 | UUGCAUAUGUAGGAUGUCCCAU |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.