| KEGG MAPK SIGNALING PATHWAY
| 267 | 205 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALLING BY NGF
| 217 | 167 | All SZGR 2.0 genes in this pathway |
| REACTOME TRIF MEDIATED TLR3 SIGNALING
| 74 | 54 | All SZGR 2.0 genes in this pathway |
| REACTOME NGF SIGNALLING VIA TRKA FROM THE PLASMA MEMBRANE
| 137 | 105 | All SZGR 2.0 genes in this pathway |
| REACTOME NUCLEAR EVENTS KINASE AND TRANSCRIPTION FACTOR ACTIVATION
| 24 | 20 | All SZGR 2.0 genes in this pathway |
| REACTOME ERKS ARE INACTIVATED
| 12 | 9 | All SZGR 2.0 genes in this pathway |
| REACTOME ERK MAPK TARGETS
| 21 | 17 | All SZGR 2.0 genes in this pathway |
| REACTOME MAP KINASE ACTIVATION IN TLR CASCADE
| 50 | 37 | All SZGR 2.0 genes in this pathway |
| REACTOME MAPK TARGETS NUCLEAR EVENTS MEDIATED BY MAP KINASES
| 30 | 24 | All SZGR 2.0 genes in this pathway |
| REACTOME TRAF6 MEDIATED INDUCTION OF NFKB AND MAP KINASES UPON TLR7 8 OR 9 ACTIVATION
| 77 | 57 | All SZGR 2.0 genes in this pathway |
| REACTOME NFKB AND MAP KINASES ACTIVATION MEDIATED BY TLR4 SIGNALING REPERTOIRE
| 72 | 53 | All SZGR 2.0 genes in this pathway |
| REACTOME MYD88 MAL CASCADE INITIATED ON PLASMA MEMBRANE
| 83 | 63 | All SZGR 2.0 genes in this pathway |
| REACTOME INNATE IMMUNE SYSTEM
| 279 | 178 | All SZGR 2.0 genes in this pathway |
| REACTOME ACTIVATED TLR4 SIGNALLING
| 93 | 69 | All SZGR 2.0 genes in this pathway |
| REACTOME IMMUNE SYSTEM
| 933 | 616 | All SZGR 2.0 genes in this pathway |
| REACTOME TOLL RECEPTOR CASCADES
| 118 | 84 | All SZGR 2.0 genes in this pathway |
| DAVICIONI MOLECULAR ARMS VS ERMS DN
| 182 | 111 | All SZGR 2.0 genes in this pathway |
| ZHOU INFLAMMATORY RESPONSE LIVE DN
| 384 | 220 | All SZGR 2.0 genes in this pathway |
| BORCZUK MALIGNANT MESOTHELIOMA UP
| 305 | 185 | All SZGR 2.0 genes in this pathway |
| GRABARCZYK BCL11B TARGETS DN
| 57 | 35 | All SZGR 2.0 genes in this pathway |
| JAEGER METASTASIS DN
| 258 | 141 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN MLL SIGNATURE 1 UP
| 380 | 236 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN MLL SIGNATURE 2 UP
| 418 | 263 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC1 TARGETS UP
| 457 | 269 | All SZGR 2.0 genes in this pathway |
| KIM WT1 TARGETS 8HR UP
| 164 | 122 | All SZGR 2.0 genes in this pathway |
| KIM WT1 TARGETS 12HR DN
| 209 | 122 | All SZGR 2.0 genes in this pathway |
| CONCANNON APOPTOSIS BY EPOXOMICIN DN
| 172 | 112 | All SZGR 2.0 genes in this pathway |
| WANG ESOPHAGUS CANCER VS NORMAL DN
| 101 | 66 | All SZGR 2.0 genes in this pathway |
| STREICHER LSM1 TARGETS DN
| 19 | 13 | All SZGR 2.0 genes in this pathway |
| SASAKI TARGETS OF TP73 AND TP63
| 11 | 7 | All SZGR 2.0 genes in this pathway |
| SIMBULAN UV RESPONSE NORMAL DN
| 33 | 27 | All SZGR 2.0 genes in this pathway |
| SIMBULAN UV RESPONSE IMMORTALIZED DN
| 31 | 26 | All SZGR 2.0 genes in this pathway |
| PUJANA CHEK2 PCC NETWORK
| 779 | 480 | All SZGR 2.0 genes in this pathway |
| GROSS HYPOXIA VIA ELK3 UP
| 209 | 139 | All SZGR 2.0 genes in this pathway |
| GROSS HYPOXIA VIA ELK3 AND HIF1A DN
| 103 | 71 | All SZGR 2.0 genes in this pathway |
| RICKMAN TUMOR DIFFERENTIATED WELL VS POORLY DN
| 382 | 224 | All SZGR 2.0 genes in this pathway |
| RICKMAN METASTASIS DN
| 261 | 155 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 6HR DN
| 514 | 330 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 48HR DN
| 428 | 306 | All SZGR 2.0 genes in this pathway |
| AMIT EGF RESPONSE 480 HELA
| 164 | 118 | All SZGR 2.0 genes in this pathway |
| TARTE PLASMA CELL VS B LYMPHOCYTE UP
| 78 | 51 | All SZGR 2.0 genes in this pathway |
| FAELT B CLL WITH VH REARRANGEMENTS UP
| 48 | 34 | All SZGR 2.0 genes in this pathway |
| CROMER METASTASIS DN
| 81 | 58 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 20HR UP
| 240 | 152 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 16HR UP
| 225 | 139 | All SZGR 2.0 genes in this pathway |
| GENTILE UV RESPONSE CLUSTER D4
| 55 | 37 | All SZGR 2.0 genes in this pathway |
| WANG SMARCE1 TARGETS DN
| 371 | 218 | All SZGR 2.0 genes in this pathway |
| GENTILE UV HIGH DOSE DN
| 312 | 203 | All SZGR 2.0 genes in this pathway |
| LEE CALORIE RESTRICTION NEOCORTEX DN
| 88 | 58 | All SZGR 2.0 genes in this pathway |
| DAZARD RESPONSE TO UV NHEK DN
| 318 | 220 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS STEM CELL AND PROGENITOR
| 681 | 420 | All SZGR 2.0 genes in this pathway |
| ZHANG GATA6 TARGETS DN
| 64 | 46 | All SZGR 2.0 genes in this pathway |
| MARZEC IL2 SIGNALING UP
| 115 | 80 | All SZGR 2.0 genes in this pathway |
| RIGGI EWING SARCOMA PROGENITOR UP
| 430 | 288 | All SZGR 2.0 genes in this pathway |
| BONOME OVARIAN CANCER SURVIVAL SUBOPTIMAL DEBULKING
| 510 | 309 | All SZGR 2.0 genes in this pathway |
| GRADE COLON VS RECTAL CANCER DN
| 56 | 36 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS UP
| 1305 | 895 | All SZGR 2.0 genes in this pathway |
| VALK AML CLUSTER 11
| 36 | 24 | All SZGR 2.0 genes in this pathway |
| GOBERT OLIGODENDROCYTE DIFFERENTIATION DN
| 1080 | 713 | All SZGR 2.0 genes in this pathway |
| VANOEVELEN MYOGENESIS SIN3A TARGETS
| 220 | 133 | All SZGR 2.0 genes in this pathway |
| PECE MAMMARY STEM CELL UP
| 146 | 75 | All SZGR 2.0 genes in this pathway |
| LIM MAMMARY STEM CELL UP
| 489 | 314 | All SZGR 2.0 genes in this pathway |
| ZWANG CLASS 1 TRANSIENTLY INDUCED BY EGF
| 516 | 308 | All SZGR 2.0 genes in this pathway |
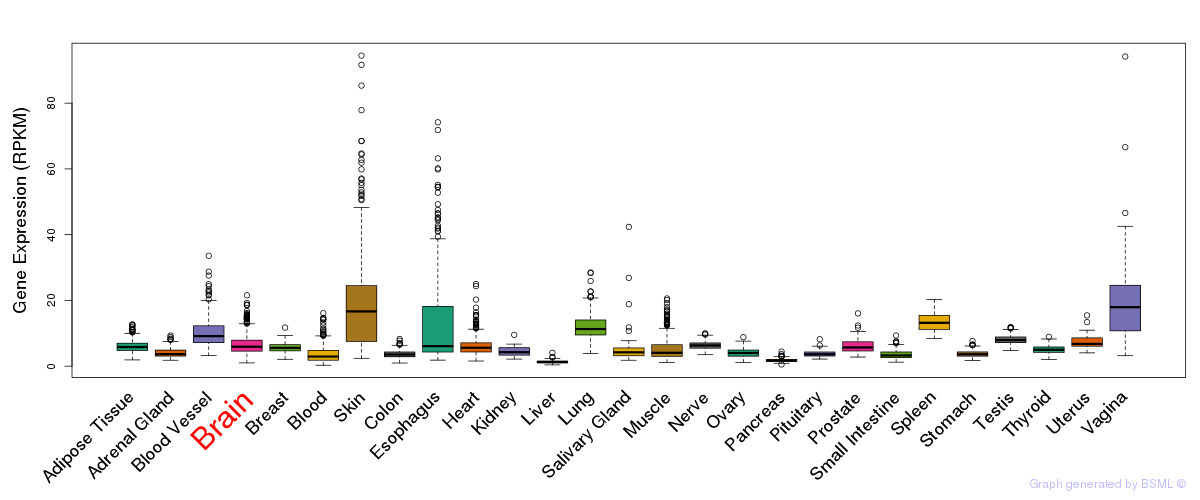
 Differentially methylated gene
Differentially methylated gene eQTL annotation
eQTL annotation