Gene Page: DYRK1A
Summary ?
| GeneID | 1859 |
| Symbol | DYRK1A |
| Synonyms | DYRK|DYRK1|HP86|MNB|MNBH|MRD7 |
| Description | dual specificity tyrosine phosphorylation regulated kinase 1A |
| Reference | MIM:600855|HGNC:HGNC:3091|Ensembl:ENSG00000157540|HPRD:09018|Vega:OTTHUMG00000086657 |
| Gene type | protein-coding |
| Map location | 21q22.13 |
| Pascal p-value | 0.049 |
| Sherlock p-value | 0.001 |
| Fetal beta | 0.656 |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 1 |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
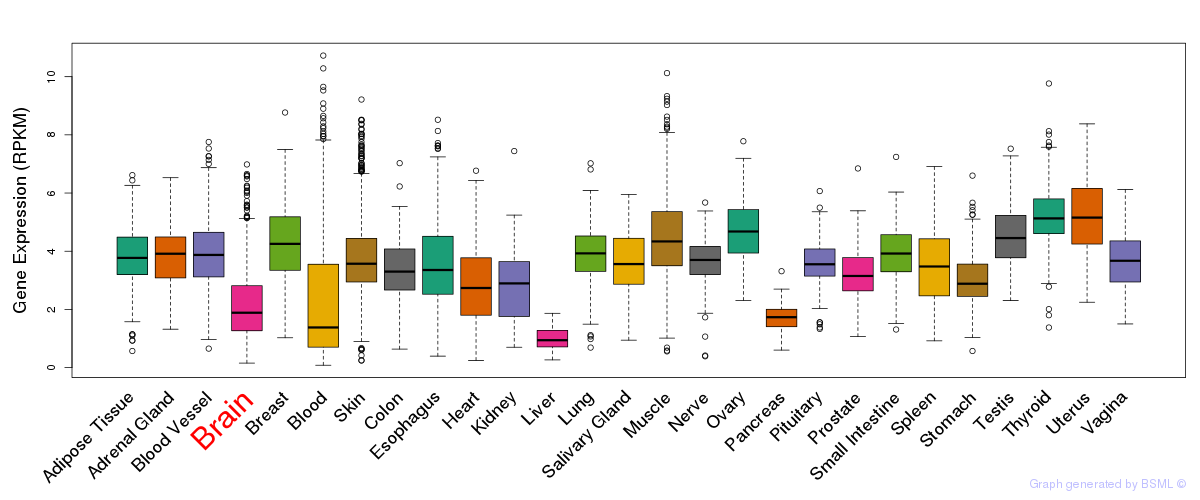
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| ANAPC1 | 0.83 | 0.87 |
| TUBGCP4 | 0.83 | 0.86 |
| ANKRD28 | 0.82 | 0.87 |
| CEP68 | 0.82 | 0.87 |
| ADAR | 0.82 | 0.87 |
| MYH10 | 0.82 | 0.84 |
| SMG7 | 0.82 | 0.87 |
| LIN54 | 0.82 | 0.85 |
| VPS13B | 0.82 | 0.84 |
| KIAA0947 | 0.82 | 0.87 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| FXYD1 | -0.66 | -0.70 |
| HIGD1B | -0.65 | -0.72 |
| MT-CO2 | -0.64 | -0.70 |
| AF347015.31 | -0.64 | -0.68 |
| AF347015.21 | -0.63 | -0.73 |
| TLCD1 | -0.62 | -0.66 |
| AC021016.1 | -0.62 | -0.65 |
| AF347015.33 | -0.61 | -0.64 |
| IFI27 | -0.61 | -0.67 |
| CST3 | -0.61 | -0.67 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0000166 | nucleotide binding | IEA | - | |
| GO:0005524 | ATP binding | IEA | - | |
| GO:0004674 | protein serine/threonine kinase activity | ISS | - | |
| GO:0004715 | non-membrane spanning protein tyrosine kinase activity | IDA | 9748265 | |
| GO:0016740 | transferase activity | IEA | - | |
| GO:0016301 | kinase activity | IEA | - | |
| GO:0043621 | protein self-association | IEA | - | |
| GO:0043621 | protein self-association | ISS | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007399 | nervous system development | TAS | neurite (GO term level: 5) | 8769099 |
| GO:0018108 | peptidyl-tyrosine phosphorylation | ISS | - | |
| GO:0046777 | protein amino acid autophosphorylation | ISS | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005634 | nucleus | ISS | - | |
| GO:0016607 | nuclear speck | ISS | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| CCNL2 | ANIA-6B | CCNM | DKFZp761A1210 | DKFZp762O195 | HCLA-ISO | HLA-ISO | PCEE | SB138 | cyclin L2 | - | HPRD,BioGRID | 14623875 |
| CREB1 | CREB | MGC9284 | cAMP responsive element binding protein 1 | DYRK1A interacts with and phosphorylates CREB. This interaction was modeled on a demonstrated interaction between human DYRK1A and CREB from rat. | BIND | 15694837 |
| DNM1 | DNM | dynamin 1 | Reconstituted Complex | BioGRID | 11877424 |
| PHYHIP | DYRK1AP3 | KIAA0273 | PAHX-AP | PAHXAP1 | phytanoyl-CoA 2-hydroxylase interacting protein | DYRK1A interacts with PAHX-AP1. | BIND | 15694837 |
| PHYHIP | DYRK1AP3 | KIAA0273 | PAHX-AP | PAHXAP1 | phytanoyl-CoA 2-hydroxylase interacting protein | - | HPRD | 15694837 |
| SFN | YWHAS | stratifin | Affinity Capture-MS | BioGRID | 15778465 |
| SFRS1 | ASF | MGC5228 | SF2 | SF2p33 | SRp30a | splicing factor, arginine/serine-rich 1 | Biochemical Activity | BioGRID | 14623875 |
| SFRS16 | CLASP | FLJ90109 | SWAP2 | splicing factor, arginine/serine-rich 16 | Biochemical Activity | BioGRID | 14623875 |
| SFRS4 | SRP75 | splicing factor, arginine/serine-rich 4 | Biochemical Activity | BioGRID | 14623875 |
| SFRS5 | HRS | SRP40 | splicing factor, arginine/serine-rich 5 | Biochemical Activity | BioGRID | 14623875 |
| WDR68 | AN11 | HAN11 | WD repeat domain 68 | Affinity Capture-Western Co-purification | BioGRID | 14593110 |
| YWHAG | 14-3-3GAMMA | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, gamma polypeptide | - | HPRD | 15324660 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| BIOCARTA SHH PATHWAY | 16 | 15 | All SZGR 2.0 genes in this pathway |
| REACTOME G0 AND EARLY G1 | 25 | 13 | All SZGR 2.0 genes in this pathway |
| REACTOME CELL CYCLE | 421 | 253 | All SZGR 2.0 genes in this pathway |
| REACTOME CELL CYCLE MITOTIC | 325 | 185 | All SZGR 2.0 genes in this pathway |
| REACTOME MITOTIC G1 G1 S PHASES | 137 | 79 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP | 1382 | 904 | All SZGR 2.0 genes in this pathway |
| WANG LMO4 TARGETS UP | 372 | 227 | All SZGR 2.0 genes in this pathway |
| TAKEDA TARGETS OF NUP98 HOXA9 FUSION 6HR DN | 41 | 29 | All SZGR 2.0 genes in this pathway |
| UDAYAKUMAR MED1 TARGETS DN | 240 | 171 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN DN | 1781 | 1082 | All SZGR 2.0 genes in this pathway |
| DARWICHE PAPILLOMA PROGRESSION RISK | 74 | 44 | All SZGR 2.0 genes in this pathway |
| SCHLOSSER SERUM RESPONSE DN | 712 | 443 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 DN | 855 | 609 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 COMMON DN | 483 | 336 | All SZGR 2.0 genes in this pathway |
| AMUNDSON RESPONSE TO ARSENITE | 217 | 143 | All SZGR 2.0 genes in this pathway |
| GEORGES TARGETS OF MIR192 AND MIR215 | 893 | 528 | All SZGR 2.0 genes in this pathway |
| SMITH TERT TARGETS UP | 145 | 91 | All SZGR 2.0 genes in this pathway |
| PENG LEUCINE DEPRIVATION DN | 187 | 122 | All SZGR 2.0 genes in this pathway |
| CHEOK RESPONSE TO HD MTX DN | 24 | 18 | All SZGR 2.0 genes in this pathway |
| PENG GLUCOSE DEPRIVATION DN | 169 | 112 | All SZGR 2.0 genes in this pathway |
| PENG GLUTAMINE DEPRIVATION DN | 337 | 230 | All SZGR 2.0 genes in this pathway |
| FLECHNER BIOPSY KIDNEY TRANSPLANT OK VS DONOR UP | 555 | 346 | All SZGR 2.0 genes in this pathway |
| CROMER METASTASIS UP | 79 | 46 | All SZGR 2.0 genes in this pathway |
| DEBIASI APOPTOSIS BY REOVIRUS INFECTION UP | 314 | 201 | All SZGR 2.0 genes in this pathway |
| DAZARD RESPONSE TO UV SCC DN | 123 | 86 | All SZGR 2.0 genes in this pathway |
| GENTILE UV RESPONSE CLUSTER D5 | 39 | 26 | All SZGR 2.0 genes in this pathway |
| GENTILE UV HIGH DOSE DN | 312 | 203 | All SZGR 2.0 genes in this pathway |
| DAZARD RESPONSE TO UV NHEK DN | 318 | 220 | All SZGR 2.0 genes in this pathway |
| BAELDE DIABETIC NEPHROPATHY DN | 434 | 302 | All SZGR 2.0 genes in this pathway |
| LEE CALORIE RESTRICTION MUSCLE DN | 51 | 28 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 STIMULATED | 1022 | 619 | All SZGR 2.0 genes in this pathway |
| FINETTI BREAST CANCERS KINOME GRAY | 15 | 10 | All SZGR 2.0 genes in this pathway |
| IWANAGA CARCINOGENESIS BY KRAS PTEN DN | 353 | 226 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS UP | 1305 | 895 | All SZGR 2.0 genes in this pathway |
| ZHAN VARIABLE EARLY DIFFERENTIATION GENES DN | 30 | 19 | All SZGR 2.0 genes in this pathway |
| YAGI AML WITH INV 16 TRANSLOCATION | 422 | 277 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 4HR DN | 254 | 158 | All SZGR 2.0 genes in this pathway |
| WIERENGA STAT5A TARGETS DN | 213 | 127 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE LATE | 1137 | 655 | All SZGR 2.0 genes in this pathway |
| PURBEY TARGETS OF CTBP1 NOT SATB1 DN | 448 | 282 | All SZGR 2.0 genes in this pathway |
| WAKABAYASHI ADIPOGENESIS PPARG BOUND 8D | 658 | 397 | All SZGR 2.0 genes in this pathway |
| LOPEZ TRANSLATION VIA FN1 SIGNALING | 35 | 21 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| let-7/98 | 1905 | 1911 | m8 | hsa-let-7abrain | UGAGGUAGUAGGUUGUAUAGUU |
| hsa-let-7bbrain | UGAGGUAGUAGGUUGUGUGGUU | ||||
| hsa-let-7cbrain | UGAGGUAGUAGGUUGUAUGGUU | ||||
| hsa-let-7dbrain | AGAGGUAGUAGGUUGCAUAGU | ||||
| hsa-let-7ebrain | UGAGGUAGGAGGUUGUAUAGU | ||||
| hsa-let-7fbrain | UGAGGUAGUAGAUUGUAUAGUU | ||||
| hsa-miR-98brain | UGAGGUAGUAAGUUGUAUUGUU | ||||
| hsa-let-7gSZ | UGAGGUAGUAGUUUGUACAGU | ||||
| hsa-let-7ibrain | UGAGGUAGUAGUUUGUGCUGU | ||||
| miR-101 | 520 | 527 | 1A,m8 | hsa-miR-101 | UACAGUACUGUGAUAACUGAAG |
| miR-129-5p | 2347 | 2353 | 1A | hsa-miR-129brain | CUUUUUGCGGUCUGGGCUUGC |
| hsa-miR-129-5p | CUUUUUGCGGUCUGGGCUUGCU | ||||
| miR-137 | 763 | 769 | m8 | hsa-miR-137 | UAUUGCUUAAGAAUACGCGUAG |
| miR-144 | 521 | 527 | 1A | hsa-miR-144 | UACAGUAUAGAUGAUGUACUAG |
| miR-145 | 1421 | 1428 | 1A,m8 | hsa-miR-145 | GUCCAGUUUUCCCAGGAAUCCCUU |
| miR-148/152 | 754 | 761 | 1A,m8 | hsa-miR-148a | UCAGUGCACUACAGAACUUUGU |
| hsa-miR-152brain | UCAGUGCAUGACAGAACUUGGG | ||||
| hsa-miR-148b | UCAGUGCAUCACAGAACUUUGU | ||||
| miR-150 | 1838 | 1845 | 1A,m8 | hsa-miR-150 | UCUCCCAACCCUUGUACCAGUG |
| miR-17-5p/20/93.mr/106/519.d | 1345 | 1351 | m8 | hsa-miR-17-5p | CAAAGUGCUUACAGUGCAGGUAGU |
| hsa-miR-20abrain | UAAAGUGCUUAUAGUGCAGGUAG | ||||
| hsa-miR-106a | AAAAGUGCUUACAGUGCAGGUAGC | ||||
| hsa-miR-106bSZ | UAAAGUGCUGACAGUGCAGAU | ||||
| hsa-miR-20bSZ | CAAAGUGCUCAUAGUGCAGGUAG | ||||
| hsa-miR-519d | CAAAGUGCCUCCCUUUAGAGUGU | ||||
| miR-182 | 330 | 336 | m8 | hsa-miR-182 | UUUGGCAAUGGUAGAACUCACA |
| miR-192/215 | 1985 | 1992 | 1A,m8 | hsa-miR-192 | CUGACCUAUGAAUUGACAGCC |
| hsa-miR-215 | AUGACCUAUGAAUUGACAGAC | ||||
| miR-193 | 785 | 791 | m8 | hsa-miR-193a | AACUGGCCUACAAAGUCCCAG |
| hsa-miR-193b | AACUGGCCCUCAAAGUCCCGCUUU | ||||
| miR-203.1 | 869 | 875 | 1A | hsa-miR-203 | UGAAAUGUUUAGGACCACUAG |
| miR-204/211 | 159 | 165 | m8 | hsa-miR-204brain | UUCCCUUUGUCAUCCUAUGCCU |
| hsa-miR-211 | UUCCCUUUGUCAUCCUUCGCCU | ||||
| miR-216 | 93 | 99 | 1A | hsa-miR-216 | UAAUCUCAGCUGGCAACUGUG |
| miR-217 | 515 | 522 | 1A,m8 | hsa-miR-217 | UACUGCAUCAGGAACUGAUUGGAU |
| miR-26 | 144 | 150 | 1A | hsa-miR-26abrain | UUCAAGUAAUCCAGGAUAGGC |
| hsa-miR-26bSZ | UUCAAGUAAUUCAGGAUAGGUU | ||||
| hsa-miR-26abrain | UUCAAGUAAUCCAGGAUAGGC | ||||
| hsa-miR-26bSZ | UUCAAGUAAUUCAGGAUAGGUU | ||||
| miR-27 | 1244 | 1250 | 1A | hsa-miR-27abrain | UUCACAGUGGCUAAGUUCCGC |
| hsa-miR-27bbrain | UUCACAGUGGCUAAGUUCUGC | ||||
| miR-30-3p | 920 | 926 | 1A | hsa-miR-30a-3p | CUUUCAGUCGGAUGUUUGCAGC |
| hsa-miR-30e-3p | CUUUCAGUCGGAUGUUUACAGC | ||||
| miR-330 | 1067 | 1074 | 1A,m8 | hsa-miR-330brain | GCAAAGCACACGGCCUGCAGAGA |
| miR-346 | 903 | 909 | 1A | hsa-miR-346brain | UGUCUGCCCGCAUGCCUGCCUCU |
| miR-363 | 69 | 75 | 1A | hsa-miR-363 | AUUGCACGGUAUCCAUCUGUAA |
| miR-378 | 2122 | 2128 | 1A | hsa-miR-378 | CUCCUGACUCCAGGUCCUGUGU |
| miR-409-5p | 1322 | 1328 | 1A | hsa-miR-409-5p | AGGUUACCCGAGCAACUUUGCA |
| miR-455 | 1308 | 1314 | 1A | hsa-miR-455 | UAUGUGCCUUUGGACUACAUCG |
| hsa-miR-455 | UAUGUGCCUUUGGACUACAUCG | ||||
| miR-485-3p | 2616 | 2622 | m8 | hsa-miR-485-3p | GUCAUACACGGCUCUCCUCUCU |
| miR-486 | 2466 | 2472 | 1A | hsa-miR-486 | UCCUGUACUGAGCUGCCCCGAG |
| miR-495 | 17 | 23 | m8 | hsa-miR-495brain | AAACAAACAUGGUGCACUUCUUU |
| miR-496 | 2688 | 2694 | 1A | hsa-miR-496 | AUUACAUGGCCAAUCUC |
| miR-544 | 1203 | 1209 | m8 | hsa-miR-544 | AUUCUGCAUUUUUAGCAAGU |
| miR-9 | 1290 | 1296 | m8 | hsa-miR-9SZ | UCUUUGGUUAUCUAGCUGUAUGA |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.