Gene Page: E2F2
Summary ?
| GeneID | 1870 |
| Symbol | E2F2 |
| Synonyms | E2F-2 |
| Description | E2F transcription factor 2 |
| Reference | MIM:600426|HGNC:HGNC:3114|Ensembl:ENSG00000007968|HPRD:02692|Vega:OTTHUMG00000003223 |
| Gene type | protein-coding |
| Map location | 1p36 |
| Pascal p-value | 0.502 |
| Fetal beta | 2.731 |
| DMG | 1 (# studies) |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg02992252 | 1 | 23857546 | E2F2 | 3.136E-4 | -0.347 | 0.04 | DMG:Wockner_2014 |
Section II. Transcriptome annotation
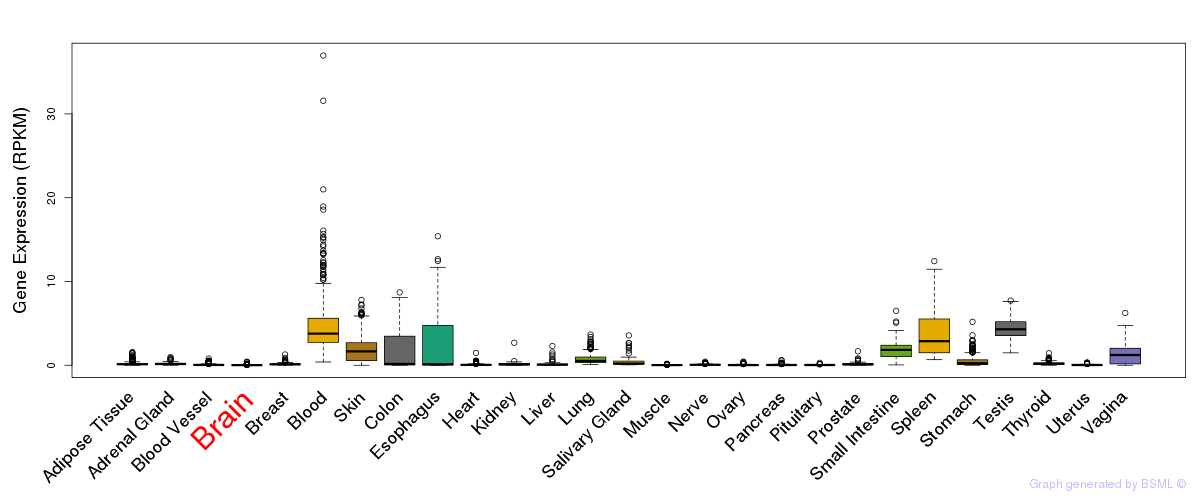
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| BCAR1 | CAS | CAS1 | CASS1 | CRKAS | P130Cas | breast cancer anti-estrogen resistance 1 | Two-hybrid | BioGRID | 16169070 |
| BRD2 | D6S113E | DKFZp686N0336 | FLJ31942 | FSH | FSRG1 | KIAA9001 | NAT | RING3 | RNF3 | bromodomain containing 2 | - | HPRD,BioGRID | 10965846 |12145330 |
| CDC25A | CDC25A2 | cell division cycle 25 homolog A (S. pombe) | E2F2 interacts with the Cdc25A promoter region. | BIND | 10766737 |
| CDC6 | CDC18L | HsCDC18 | HsCDC6 | cell division cycle 6 homolog (S. cerevisiae) | E2F2 interacts with the Cdc6 promoter region. | BIND | 10766737 |
| CDK3 | - | cyclin-dependent kinase 3 | - | HPRD,BioGRID | 8846921 |11733001 |
| E2F1 | E2F-1 | RBAP1 | RBBP3 | RBP3 | E2F transcription factor 1 | E2F2 interacts with the E2F1 promoter region. | BIND | 10766737 |
| E2F4 | E2F-4 | E2F transcription factor 4, p107/p130-binding | E2F4 interacts with the E2F2 promoter. | BIND | 15861133 |
| EAPP | BM036 | C14orf11 | FLJ20578 | MGC4957 | E2F-associated phosphoprotein | E2F2 interacts with EAPP. | BIND | 15716352 |
| FHL2 | AAG11 | DRAL | SLIM3 | four and a half LIM domains 2 | Two-hybrid | BioGRID | 12411495 |
| FHL2 | AAG11 | DRAL | SLIM3 | four and a half LIM domains 2 | E2F2 interacts with a molecule expressed by Gene FHL2. The precise molecular variant of FHL2 involved in this interaction is not specified. | BIND | 12411495 |
| GAB2 | KIAA0571 | GRB2-associated binding protein 2 | The Gab2 promoter region interacts with E2F2. | BIND | 15574337 |
| JMY | FLJ37870 | MGC163496 | junction-mediating and regulatory protein | E2F2 interacts with the JMY promoter. | BIND | 15706352 |
| MT1G | MGC12386 | MT1 | MT1K | metallothionein 1G | E2F2 interacts with the MT1G promoter. | BIND | 15735762 |
| MYBL2 | B-MYB | BMYB | MGC15600 | v-myb myeloblastosis viral oncogene homolog (avian)-like 2 | E2F2 interacts with the B-myb promoter region. | BIND | 10766737 |
| PPP1R13B | ASPP1 | KIAA0771 | p53BP2-like | p85 | protein phosphatase 1, regulatory (inhibitor) subunit 13B | E2F2 interacts with the PPP1R13B (ASPP1) promoter. | BIND | 15706352 |
| RB1 | OSRC | RB | p105-Rb | pRb | pp110 | retinoblastoma 1 | E2F-2 interacts with pRb. | BIND | 8246996 |
| RB1 | OSRC | RB | p105-Rb | pRb | pp110 | retinoblastoma 1 | - | HPRD,BioGRID | 12502741 |
| RB1 | OSRC | RB | p105-Rb | pRb | pp110 | retinoblastoma 1 | E2F1 interacts with pRb. | BIND | 8246996 |
| RB1 | OSRC | RB | p105-Rb | pRb | pp110 | retinoblastoma 1 | pRB interacts with E2F-2. | BIND | 15949438 |
| RBL1 | CP107 | MGC40006 | PRB1 | p107 | retinoblastoma-like 1 (p107) | E2F2 interacts with the p107 promoter region. | BIND | 10766737 |
| RBL2 | FLJ26459 | P130 | Rb2 | retinoblastoma-like 2 (p130) | RBL2 (p130) interacts with the E2F2 promoter. | BIND | 15861133 |
| RNF144A | KIAA0161 | RNF144 | UBCE7IP4 | ring finger protein 144A | Two-hybrid | BioGRID | 12411495 |
| RYBP | AAP1 | DEDAF | YEAF1 | RING1 and YY1 binding protein | E2F2 interacts with RYBP. | BIND | 12411495 |
| RYBP | AAP1 | DEDAF | YEAF1 | RING1 and YY1 binding protein | Affinity Capture-Western Two-hybrid | BioGRID | 12411495 |
| SETD8 | KMT5A | PR-Set7 | SET07 | SET8 | SET domain containing (lysine methyltransferase) 8 | E2F2 interacts with SET 07. | BIND | 12411495 |
| SETD8 | KMT5A | PR-Set7 | SET07 | SET8 | SET domain containing (lysine methyltransferase) 8 | Two-hybrid | BioGRID | 12411495 |
| SP1 | - | Sp1 transcription factor | - | HPRD | 8657141 |8657142 |9694791 |
| SPIB | SPI-B | Spi-B transcription factor (Spi-1/PU.1 related) | Two-hybrid | BioGRID | 12748276 |
| TFDP1 | DP1 | DRTF1 | Dp-1 | transcription factor Dp-1 | - | HPRD | 7739537 |8755520 |
| TFDP2 | DP2 | Dp-2 | transcription factor Dp-2 (E2F dimerization partner 2) | - | HPRD | 7739537 |8755520 |
| TP53BP2 | 53BP2 | ASPP2 | BBP | PPP1R13A | p53BP2 | tumor protein p53 binding protein, 2 | E2F2 interacts with the TP53BP2 (ASPP2) promoter. | BIND | 15706352 |
| TP53INP1 | DKFZp434M1317 | FLJ22139 | SIP | TP53DINP1 | TP53INP1A | TP53INP1B | Teap | p53DINP1 | tumor protein p53 inducible nuclear protein 1 | E2F2 interacts with the TP53INP1 promoter. | BIND | 15706352 |
| UXT | ART-27 | ubiquitously-expressed transcript | E2F2 interacts with the UXT chromatin. | BIND | 11799066 |
| YY1 | DELTA | INO80S | NF-E1 | UCRBP | YIN-YANG-1 | YY1 transcription factor | - | HPRD | 12411495 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG CELL CYCLE | 128 | 84 | All SZGR 2.0 genes in this pathway |
| KEGG PATHWAYS IN CANCER | 328 | 259 | All SZGR 2.0 genes in this pathway |
| KEGG PANCREATIC CANCER | 70 | 56 | All SZGR 2.0 genes in this pathway |
| KEGG GLIOMA | 65 | 56 | All SZGR 2.0 genes in this pathway |
| KEGG PROSTATE CANCER | 89 | 75 | All SZGR 2.0 genes in this pathway |
| KEGG MELANOMA | 71 | 57 | All SZGR 2.0 genes in this pathway |
| KEGG BLADDER CANCER | 42 | 33 | All SZGR 2.0 genes in this pathway |
| KEGG CHRONIC MYELOID LEUKEMIA | 73 | 59 | All SZGR 2.0 genes in this pathway |
| KEGG SMALL CELL LUNG CANCER | 84 | 67 | All SZGR 2.0 genes in this pathway |
| KEGG NON SMALL CELL LUNG CANCER | 54 | 47 | All SZGR 2.0 genes in this pathway |
| SA G1 AND S PHASES | 15 | 10 | All SZGR 2.0 genes in this pathway |
| SA REG CASCADE OF CYCLIN EXPR | 13 | 7 | All SZGR 2.0 genes in this pathway |
| PID E2F PATHWAY | 74 | 48 | All SZGR 2.0 genes in this pathway |
| PID P53 DOWNSTREAM PATHWAY | 137 | 94 | All SZGR 2.0 genes in this pathway |
| PID RB 1PATHWAY | 65 | 46 | All SZGR 2.0 genes in this pathway |
| REACTOME CELL CYCLE | 421 | 253 | All SZGR 2.0 genes in this pathway |
| REACTOME CELL CYCLE MITOTIC | 325 | 185 | All SZGR 2.0 genes in this pathway |
| REACTOME G1 PHASE | 38 | 23 | All SZGR 2.0 genes in this pathway |
| REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | 11 | 5 | All SZGR 2.0 genes in this pathway |
| REACTOME M G1 TRANSITION | 81 | 45 | All SZGR 2.0 genes in this pathway |
| REACTOME MITOTIC G1 G1 S PHASES | 137 | 79 | All SZGR 2.0 genes in this pathway |
| REACTOME MITOTIC M M G1 PHASES | 172 | 98 | All SZGR 2.0 genes in this pathway |
| REACTOME ASSEMBLY OF THE PRE REPLICATIVE COMPLEX | 65 | 36 | All SZGR 2.0 genes in this pathway |
| REACTOME DNA REPLICATION | 192 | 110 | All SZGR 2.0 genes in this pathway |
| GAL LEUKEMIC STEM CELL DN | 244 | 153 | All SZGR 2.0 genes in this pathway |
| WANG CLIM2 TARGETS UP | 269 | 146 | All SZGR 2.0 genes in this pathway |
| KINSEY TARGETS OF EWSR1 FLII FUSION UP | 1278 | 748 | All SZGR 2.0 genes in this pathway |
| SCHLOSSER SERUM RESPONSE DN | 712 | 443 | All SZGR 2.0 genes in this pathway |
| PATIL LIVER CANCER | 747 | 453 | All SZGR 2.0 genes in this pathway |
| NUYTTEN EZH2 TARGETS DN | 1024 | 594 | All SZGR 2.0 genes in this pathway |
| GARCIA TARGETS OF FLI1 AND DAX1 DN | 176 | 104 | All SZGR 2.0 genes in this pathway |
| PASQUALUCCI LYMPHOMA BY GC STAGE DN | 165 | 104 | All SZGR 2.0 genes in this pathway |
| ZHANG RESPONSE TO IKK INHIBITOR AND TNF DN | 103 | 64 | All SZGR 2.0 genes in this pathway |
| RADAEVA RESPONSE TO IFNA1 UP | 52 | 40 | All SZGR 2.0 genes in this pathway |
| REN BOUND BY E2F | 61 | 40 | All SZGR 2.0 genes in this pathway |
| VERNELL RETINOBLASTOMA PATHWAY UP | 70 | 47 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS MATURE CELL | 293 | 160 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 UNSTIMULATED | 1229 | 713 | All SZGR 2.0 genes in this pathway |
| ZHANG GATA6 TARGETS DN | 64 | 46 | All SZGR 2.0 genes in this pathway |
| HELLER SILENCED BY METHYLATION DN | 105 | 67 | All SZGR 2.0 genes in this pathway |
| HELLER HDAC TARGETS SILENCED BY METHYLATION DN | 281 | 179 | All SZGR 2.0 genes in this pathway |
| AGUIRRE PANCREATIC CANCER COPY NUMBER DN | 238 | 145 | All SZGR 2.0 genes in this pathway |
| LEE EARLY T LYMPHOCYTE UP | 107 | 59 | All SZGR 2.0 genes in this pathway |
| MEISSNER BRAIN HCP WITH H3K4ME3 AND H3K27ME3 | 1069 | 729 | All SZGR 2.0 genes in this pathway |
| MILI PSEUDOPODIA HAPTOTAXIS DN | 668 | 419 | All SZGR 2.0 genes in this pathway |
| WHITFIELD CELL CYCLE G1 S | 147 | 76 | All SZGR 2.0 genes in this pathway |
| WANG RESPONSE TO GSK3 INHIBITOR SB216763 DN | 374 | 217 | All SZGR 2.0 genes in this pathway |
| DUTERTRE ESTRADIOL RESPONSE 24HR UP | 324 | 193 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS DN | 1972 | 1213 | All SZGR 2.0 genes in this pathway |
| BHAT ESR1 TARGETS NOT VIA AKT1 UP | 211 | 131 | All SZGR 2.0 genes in this pathway |
| BHAT ESR1 TARGETS VIA AKT1 UP | 281 | 183 | All SZGR 2.0 genes in this pathway |
| GOBERT OLIGODENDROCYTE DIFFERENTIATION UP | 570 | 339 | All SZGR 2.0 genes in this pathway |
| HUANG GATA2 TARGETS DN | 72 | 52 | All SZGR 2.0 genes in this pathway |
| LE NEURONAL DIFFERENTIATION DN | 19 | 16 | All SZGR 2.0 genes in this pathway |