Gene Page: EEF1D
Summary ?
| GeneID | 1936 |
| Symbol | EEF1D |
| Synonyms | EF-1D|EF1D|FP1047 |
| Description | eukaryotic translation elongation factor 1 delta |
| Reference | MIM:130592|HGNC:HGNC:3211|Ensembl:ENSG00000104529|HPRD:00560|Vega:OTTHUMG00000165191 |
| Gene type | protein-coding |
| Map location | 8q24.3 |
| Pascal p-value | 0.016 |
| Sherlock p-value | 0.079 |
| Fetal beta | 0.894 |
| DMG | 1 (# studies) |
| eGene | Myers' cis & trans |
| Support | CompositeSet |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Montano_2016 | Genome-wide DNA methylation analysis | This dataset includes 172 replicated associations between CpGs with schizophrenia. | 2 |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 2 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg17636309 | 8 | 144672242 | EEF1D | 2.06E-4 | 0.007 | 0.158 | DMG:Montano_2016 |
| cg04249066 | 8 | 144680433 | EEF1D;TIGD5 | 4.109E-4 | -0.314 | 0.044 | DMG:Wockner_2014 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs924274 | chr1 | 61479290 | EEF1D | 1936 | 0.11 | trans | ||
| rs4436485 | chr10 | 103245360 | EEF1D | 1936 | 0.03 | trans |
Section II. Transcriptome annotation
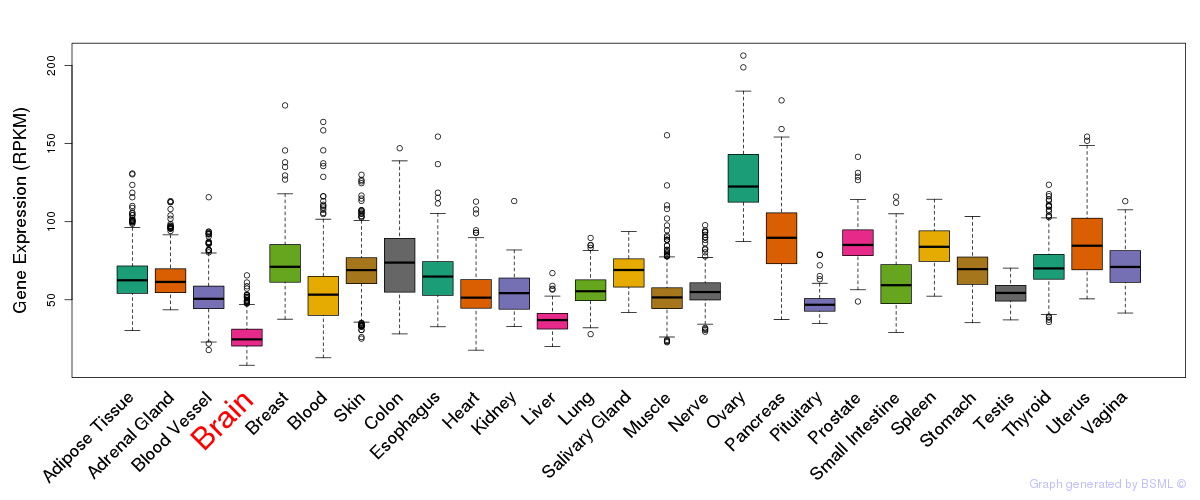
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| PCDHGC4 | 0.90 | 0.89 |
| SARM1 | 0.89 | 0.95 |
| MAGED1 | 0.89 | 0.92 |
| TBCD | 0.89 | 0.92 |
| FGD1 | 0.88 | 0.94 |
| PACS1 | 0.88 | 0.94 |
| DTX1 | 0.88 | 0.92 |
| PIK3R2 | 0.88 | 0.94 |
| SMPD3 | 0.88 | 0.94 |
| WDR40C | 0.88 | 0.92 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| MT-CO2 | -0.75 | -0.86 |
| AF347015.31 | -0.75 | -0.84 |
| AF347015.33 | -0.74 | -0.83 |
| AF347015.27 | -0.74 | -0.84 |
| AF347015.8 | -0.72 | -0.85 |
| MT-CYB | -0.72 | -0.83 |
| S100B | -0.71 | -0.78 |
| AF347015.15 | -0.70 | -0.82 |
| AF347015.2 | -0.70 | -0.84 |
| FXYD1 | -0.70 | -0.78 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ASCC2 | ASC1p100 | activating signal cointegrator 1 complex subunit 2 | Two-hybrid | BioGRID | 16169070 |
| C10orf4 | F26C11.1-like | FRA10A | FRA10AC1 | chromosome 10 open reading frame 4 | Two-hybrid | BioGRID | 16169070 |
| CD48 | BCM1 | BLAST | BLAST1 | MEM-102 | SLAMF2 | hCD48 | mCD48 | CD48 molecule | Two-hybrid | BioGRID | 16169070 |
| CDC2 | CDC28A | CDK1 | DKFZp686L20222 | MGC111195 | cell division cycle 2, G1 to S and G2 to M | - | HPRD | 8051108 |
| CRMP1 | DPYSL1 | DRP-1 | DRP1 | collapsin response mediator protein 1 | Two-hybrid | BioGRID | 16169070 |
| CSNK2A1 | CK2A1 | CKII | casein kinase 2, alpha 1 polypeptide | - | HPRD,BioGRID | 10094407 |
| DARS | DKFZp781B11202 | MGC111579 | aspartyl-tRNA synthetase | DRS interacts with unspecified isoform of EF-1-delta. | BIND | 11829477 |
| EEF1A2 | EEF1AL | EF-1-alpha-2 | EF1A | FLJ41696 | HS1 | STN | STNL | eukaryotic translation elongation factor 1 alpha 2 | Affinity Capture-MS | BioGRID | 17353931 |
| EEF1D | EF-1D | EF1D | FLJ20897 | FP1047 | eukaryotic translation elongation factor 1 delta (guanine nucleotide exchange protein) | - | HPRD | 10094407 |
| EEF1D | EF-1D | EF1D | FLJ20897 | FP1047 | eukaryotic translation elongation factor 1 delta (guanine nucleotide exchange protein) | Two-hybrid | BioGRID | 16169070 |
| EEF1G | EF1G | GIG35 | eukaryotic translation elongation factor 1 gamma | Two-hybrid | BioGRID | 16169070 |16189514 |
| ELF3 | EPR-1 | ERT | ESE-1 | ESX | E74-like factor 3 (ets domain transcription factor, epithelial-specific ) | Affinity Capture-MS | BioGRID | 17353931 |
| FARSA | CML33 | FARSL | FARSLA | FRSA | PheHA | phenylalanyl-tRNA synthetase, alpha subunit | FRS-alpha interacts with an unspecified isoform of EF-1-delta. | BIND | 11829477 |
| GARS | CMT2D | DSMAV | GlyRS | HMN5 | SMAD1 | glycyl-tRNA synthetase | GRS interacts with an unspecified isoform of EF-1-delta. | BIND | 11829477 |
| GARS | CMT2D | DSMAV | GlyRS | HMN5 | SMAD1 | glycyl-tRNA synthetase | - | HPRD,BioGRID | 11829477 |
| HEXDC | FLJ23825 | hexosaminidase (glycosyl hydrolase family 20, catalytic domain) containing | Two-hybrid | BioGRID | 16169070 |
| KTN1 | CG1 | KIAA0004 | KNT | MGC133337 | MU-RMS-40.19 | kinectin 1 (kinesin receptor) | - | HPRD,BioGRID | 12773547 |
| RIPK3 | RIP3 | receptor-interacting serine-threonine kinase 3 | - | HPRD | 14743216 |
| TPT1 | FLJ27337 | HRF | TCTP | p02 | tumor protein, translationally-controlled 1 | - | HPRD | 14623968 |15062873 |
| VARS | G7A | VARS1 | VARS2 | valyl-tRNA synthetase | - | HPRD | 2556394 |3169261 |
| VARS | G7A | VARS1 | VARS2 | valyl-tRNA synthetase | - | HPRD,BioGRID | 2556394 |3169261|8294461 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| REACTOME TRANSLATION | 222 | 75 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF PROTEINS | 518 | 242 | All SZGR 2.0 genes in this pathway |
| HOLLMANN APOPTOSIS VIA CD40 DN | 267 | 178 | All SZGR 2.0 genes in this pathway |
| GAZDA DIAMOND BLACKFAN ANEMIA ERYTHROID DN | 493 | 298 | All SZGR 2.0 genes in this pathway |
| DAVICIONI MOLECULAR ARMS VS ERMS DN | 182 | 111 | All SZGR 2.0 genes in this pathway |
| CASORELLI APL SECONDARY VS DE NOVO DN | 9 | 5 | All SZGR 2.0 genes in this pathway |
| OSWALD HEMATOPOIETIC STEM CELL IN COLLAGEN GEL UP | 233 | 161 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN NPM1 MUTATED SIGNATURE 1 DN | 126 | 86 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN NPM1 MUTATED SIGNATURE 2 DN | 77 | 46 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN MLL SIGNATURE 1 DN | 242 | 165 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN MLL SIGNATURE 2 DN | 281 | 186 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN NPM1 SIGNATURE 3 DN | 162 | 116 | All SZGR 2.0 genes in this pathway |
| CHOW RASSF1 TARGETS DN | 29 | 19 | All SZGR 2.0 genes in this pathway |
| DARWICHE SKIN TUMOR PROMOTER DN | 185 | 115 | All SZGR 2.0 genes in this pathway |
| DARWICHE PAPILLOMA RISK LOW UP | 162 | 104 | All SZGR 2.0 genes in this pathway |
| DARWICHE PAPILLOMA RISK HIGH UP | 147 | 101 | All SZGR 2.0 genes in this pathway |
| DARWICHE SQUAMOUS CELL CARCINOMA UP | 146 | 104 | All SZGR 2.0 genes in this pathway |
| LI AMPLIFIED IN LUNG CANCER | 178 | 108 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 UP | 309 | 199 | All SZGR 2.0 genes in this pathway |
| LOPEZ MBD TARGETS | 957 | 597 | All SZGR 2.0 genes in this pathway |
| ROPERO HDAC2 TARGETS | 114 | 71 | All SZGR 2.0 genes in this pathway |
| RICKMAN TUMOR DIFFERENTIATED WELL VS POORLY UP | 236 | 139 | All SZGR 2.0 genes in this pathway |
| BENPORATH EED TARGETS | 1062 | 725 | All SZGR 2.0 genes in this pathway |
| NIKOLSKY BREAST CANCER 8Q23 Q24 AMPLICON | 157 | 87 | All SZGR 2.0 genes in this pathway |
| FLECHNER BIOPSY KIDNEY TRANSPLANT OK VS DONOR UP | 555 | 346 | All SZGR 2.0 genes in this pathway |
| LEI MYB TARGETS | 318 | 215 | All SZGR 2.0 genes in this pathway |
| FERRANDO T ALL WITH MLL ENL FUSION DN | 87 | 57 | All SZGR 2.0 genes in this pathway |
| TAKAO RESPONSE TO UVB RADIATION UP | 86 | 55 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS LATE PROGENITOR | 544 | 307 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| ZHANG RESPONSE TO CANTHARIDIN DN | 69 | 46 | All SZGR 2.0 genes in this pathway |
| LEE CALORIE RESTRICTION NEOCORTEX DN | 88 | 58 | All SZGR 2.0 genes in this pathway |
| DAIRKEE CANCER PRONE RESPONSE BPA E2 | 118 | 65 | All SZGR 2.0 genes in this pathway |
| GRADE COLON CANCER UP | 871 | 505 | All SZGR 2.0 genes in this pathway |
| WEST ADRENOCORTICAL TUMOR DN | 546 | 362 | All SZGR 2.0 genes in this pathway |
| VANTVEER BREAST CANCER BRCA1 UP | 36 | 18 | All SZGR 2.0 genes in this pathway |
| AGUIRRE PANCREATIC CANCER COPY NUMBER UP | 298 | 174 | All SZGR 2.0 genes in this pathway |
| SWEET LUNG CANCER KRAS UP | 491 | 316 | All SZGR 2.0 genes in this pathway |
| SWEET KRAS ONCOGENIC SIGNATURE | 89 | 56 | All SZGR 2.0 genes in this pathway |
| HSIAO HOUSEKEEPING GENES | 389 | 245 | All SZGR 2.0 genes in this pathway |
| CAIRO HEPATOBLASTOMA CLASSES UP | 605 | 377 | All SZGR 2.0 genes in this pathway |
| LI DCP2 BOUND MRNA | 89 | 57 | All SZGR 2.0 genes in this pathway |
| JOHNSTONE PARVB TARGETS 3 DN | 918 | 550 | All SZGR 2.0 genes in this pathway |
| FEVR CTNNB1 TARGETS DN | 553 | 343 | All SZGR 2.0 genes in this pathway |
| RATTENBACHER BOUND BY CELF1 | 467 | 251 | All SZGR 2.0 genes in this pathway |
| PECE MAMMARY STEM CELL UP | 146 | 75 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 1ST EGF PULSE ONLY | 1839 | 928 | All SZGR 2.0 genes in this pathway |
| ZWANG EGF INTERVAL UP | 105 | 46 | All SZGR 2.0 genes in this pathway |