Gene Page: EIF4E
Summary ?
| GeneID | 1977 |
| Symbol | EIF4E |
| Synonyms | AUTS19|CBP|EIF4E1|EIF4EL1|EIF4F|eIF-4E |
| Description | eukaryotic translation initiation factor 4E |
| Reference | MIM:133440|HGNC:HGNC:3287|HPRD:00591| |
| Gene type | protein-coding |
| Map location | 4q23 |
| Pascal p-value | 0.511 |
| Sherlock p-value | 0.315 |
| Fetal beta | 0.776 |
| DMG | 1 (# studies) |
| eGene | Myers' cis & trans Meta |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 3 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.0067 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg11037477 | 4 | 99851008 | EIF4E | 1.465E-4 | 0.563 | 0.031 | DMG:Wockner_2014 |
| cg15633390 | 4 | 99851211 | EIF4E | 3.028E-4 | 0.657 | 0.04 | DMG:Wockner_2014 |
| cg14972143 | 4 | 99851003 | EIF4E | 3.611E-4 | 0.664 | 0.042 | DMG:Wockner_2014 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs4729109 | chr7 | 93760673 | EIF4E | 1977 | 0.18 | trans | ||
| rs7035652 | chr9 | 117779879 | EIF4E | 1977 | 0.08 | trans |
Section II. Transcriptome annotation
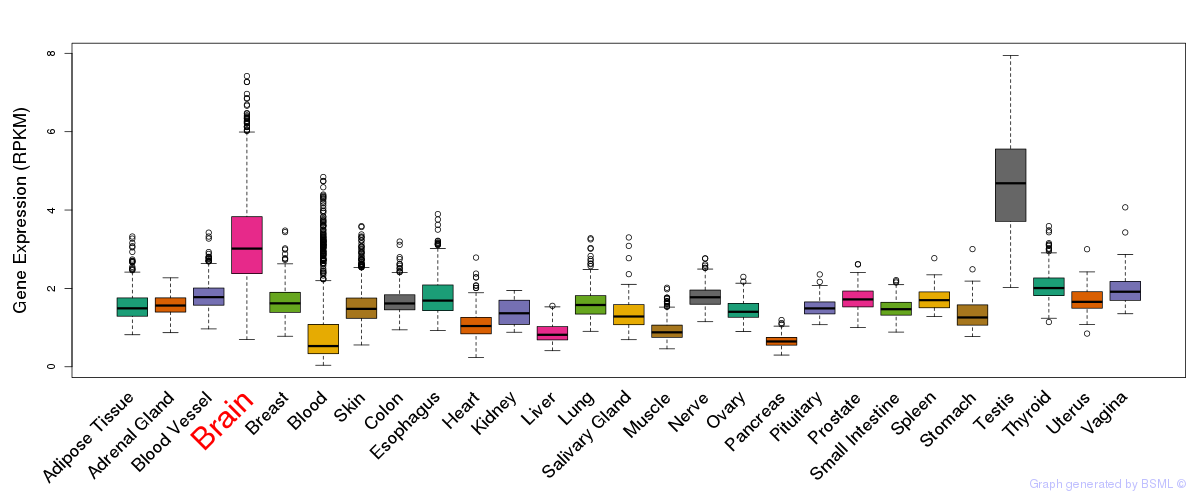
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AP003117.1 | 0.79 | 0.84 |
| CTBS | 0.77 | 0.73 |
| CRYL1 | 0.77 | 0.76 |
| TMBIM1 | 0.77 | 0.81 |
| SERPINB6 | 0.76 | 0.78 |
| GRAMD3 | 0.75 | 0.81 |
| LACTB2 | 0.75 | 0.78 |
| CD9 | 0.74 | 0.80 |
| PIR | 0.74 | 0.75 |
| CSRP1 | 0.74 | 0.79 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| KIAA1543 | -0.60 | -0.64 |
| NOVA2 | -0.60 | -0.64 |
| CHD3 | -0.60 | -0.63 |
| DPF1 | -0.59 | -0.64 |
| MARCH4 | -0.59 | -0.63 |
| ZNF821 | -0.59 | -0.63 |
| KIAA1949 | -0.58 | -0.60 |
| MPP3 | -0.58 | -0.61 |
| GPSM1 | -0.58 | -0.60 |
| KIF21B | -0.58 | -0.62 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0000339 | RNA cap binding | TAS | 3469651 | |
| GO:0003743 | translation initiation factor activity | ISS | - | |
| GO:0005515 | protein binding | IPI | 9878069 |10856257 |15153109 |15247416 |16739988 |17353931 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006413 | translational initiation | IEA | - | |
| GO:0006417 | regulation of translation | IEA | - | |
| GO:0044419 | interspecies interaction between organisms | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005829 | cytosol | EXP | 7935836 |8449919 |12588972 |15809305 | |
| GO:0005737 | cytoplasm | IEA | - | |
| GO:0016281 | eukaryotic translation initiation factor 4F complex | TAS | 3469651 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| EIF3B | EIF3-ETA | EIF3-P110 | EIF3-P116 | EIF3S9 | MGC104664 | MGC131875 | PRT1 | eukaryotic translation initiation factor 3, subunit B | Affinity Capture-Western | BioGRID | 16541103 |
| EIF4A1 | DDX2A | EIF-4A | EIF4A | eukaryotic translation initiation factor 4A, isoform 1 | Affinity Capture-MS Affinity Capture-Western | BioGRID | 16648488 |17353931 |
| EIF4A2 | BM-010 | DDX2B | EIF4A | EIF4F | eukaryotic translation initiation factor 4A, isoform 2 | Affinity Capture-MS | BioGRID | 17353931 |
| EIF4E | CBP | EIF4E1 | EIF4EL1 | EIF4F | MGC111573 | eukaryotic translation initiation factor 4E | Affinity Capture-Western | BioGRID | 16648488 |
| EIF4EBP1 | 4E-BP1 | 4EBP1 | BP-1 | MGC4316 | PHAS-I | eukaryotic translation initiation factor 4E binding protein 1 | EIF4E interacts with EIF4EBP1 (4EBP1). | BIND | 10022874 |
| EIF4EBP1 | 4E-BP1 | 4EBP1 | BP-1 | MGC4316 | PHAS-I | eukaryotic translation initiation factor 4E binding protein 1 | - | HPRD,BioGRID | 11605658 |
| EIF4EBP2 | 4EBP2 | PHASII | eukaryotic translation initiation factor 4E binding protein 2 | - | HPRD,BioGRID | 7651417 |7935836 |
| EIF4EBP3 | 4E-BP3 | eukaryotic translation initiation factor 4E binding protein 3 | - | HPRD,BioGRID | 12482586 |
| EIF4ENIF1 | 4E-T | Clast4 | FLJ21601 | FLJ26551 | eukaryotic translation initiation factor 4E nuclear import factor 1 | - | HPRD,BioGRID | 10856257 |
| EIF4G1 | DKFZp686A1451 | EIF4F | EIF4G | p220 | eukaryotic translation initiation factor 4 gamma, 1 | - | HPRD,BioGRID | 10600798 |
| EIF4G1 | DKFZp686A1451 | EIF4F | EIF4G | p220 | eukaryotic translation initiation factor 4 gamma, 1 | - | HPRD | 9372926 |10600798|10600798 |
| EIF4G1 | DKFZp686A1451 | EIF4F | EIF4G | p220 | eukaryotic translation initiation factor 4 gamma, 1 | EIF4E interacts with eIF4G1. This interaction was modeled on a demonstrated interaction between mouse eIF4E and human eIF4G1. | BIND | 9878069 |
| EIF4G1 | DKFZp686A1451 | EIF4F | EIF4G | p220 | eukaryotic translation initiation factor 4 gamma, 1 | eIF4GI interacts with eIF4E. This interaction was modeled on a demonstrated interaction between human eIF4GI and monkey eIF4E. | BIND | 15361857 |
| EIF4G1 | DKFZp686A1451 | EIF4F | EIF4G | p220 | eukaryotic translation initiation factor 4 gamma, 1 | eIF-4E interacts with an unspecified isoform of eIF-4G. | BIND | 15897904 |
| EIF4G2 | AAG1 | DAP5 | FLJ41344 | NAT1 | p97 | eukaryotic translation initiation factor 4 gamma, 2 | Affinity Capture-Western Far Western | BioGRID | 9418880 |
| EIF4G2 | AAG1 | DAP5 | FLJ41344 | NAT1 | p97 | eukaryotic translation initiation factor 4 gamma, 2 | - | HPRD | 10394359 |
| EIF4G3 | eIF4GII | eukaryotic translation initiation factor 4 gamma, 3 | - | HPRD | 9418880 |
| MAPKAPK5 | PRAK | mitogen-activated protein kinase-activated protein kinase 5 | - | HPRD | 9628874 |
| MKNK1 | MNK1 | MAP kinase interacting serine/threonine kinase 1 | Affinity Capture-Western | BioGRID | 9878069 |
| MKNK1 | MNK1 | MAP kinase interacting serine/threonine kinase 1 | EIF4E interacts with Mnk1. This interaction was modeled on a demonstrated interaction between mouse eIF4E and human Mnk1. | BIND | 9878069 |
| PML | MYL | PP8675 | RNF71 | TRIM19 | promyelocytic leukemia | - | HPRD,BioGRID | 11500381 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG MTOR SIGNALING PATHWAY | 52 | 40 | All SZGR 2.0 genes in this pathway |
| KEGG INSULIN SIGNALING PATHWAY | 137 | 103 | All SZGR 2.0 genes in this pathway |
| BIOCARTA EIF PATHWAY | 16 | 9 | All SZGR 2.0 genes in this pathway |
| BIOCARTA MTOR PATHWAY | 23 | 15 | All SZGR 2.0 genes in this pathway |
| BIOCARTA EIF4 PATHWAY | 24 | 19 | All SZGR 2.0 genes in this pathway |
| BIOCARTA IGF1MTOR PATHWAY | 20 | 14 | All SZGR 2.0 genes in this pathway |
| BIOCARTA SARS PATHWAY | 10 | 5 | All SZGR 2.0 genes in this pathway |
| ST ERK1 ERK2 MAPK PATHWAY | 32 | 25 | All SZGR 2.0 genes in this pathway |
| ST P38 MAPK PATHWAY | 37 | 28 | All SZGR 2.0 genes in this pathway |
| PID MET PATHWAY | 80 | 60 | All SZGR 2.0 genes in this pathway |
| PID MYC ACTIV PATHWAY | 79 | 62 | All SZGR 2.0 genes in this pathway |
| PID MTOR 4PATHWAY | 69 | 55 | All SZGR 2.0 genes in this pathway |
| PID P38 ALPHA BETA DOWNSTREAM PATHWAY | 38 | 29 | All SZGR 2.0 genes in this pathway |
| REACTOME TRANSLATION | 222 | 75 | All SZGR 2.0 genes in this pathway |
| REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | 66 | 45 | All SZGR 2.0 genes in this pathway |
| REACTOME INSULIN RECEPTOR SIGNALLING CASCADE | 87 | 64 | All SZGR 2.0 genes in this pathway |
| REACTOME PROCESSING OF CAPPED INTRON CONTAINING PRE MRNA | 140 | 77 | All SZGR 2.0 genes in this pathway |
| REACTOME ACTIVATION OF THE MRNA UPON BINDING OF THE CAP BINDING COMPLEX AND EIFS AND SUBSEQUENT BINDING TO 43S | 84 | 30 | All SZGR 2.0 genes in this pathway |
| REACTOME TRANSPORT OF MATURE TRANSCRIPT TO CYTOPLASM | 54 | 34 | All SZGR 2.0 genes in this pathway |
| REACTOME MRNA PROCESSING | 161 | 86 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF PROTEINS | 518 | 242 | All SZGR 2.0 genes in this pathway |
| REACTOME 3 UTR MEDIATED TRANSLATIONAL REGULATION | 176 | 51 | All SZGR 2.0 genes in this pathway |
| REACTOME TRANSPORT OF MATURE MRNA DERIVED FROM AN INTRONLESS TRANSCRIPT | 33 | 21 | All SZGR 2.0 genes in this pathway |
| REACTOME DEADENYLATION OF MRNA | 22 | 14 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF MRNA | 284 | 128 | All SZGR 2.0 genes in this pathway |
| REACTOME DEADENYLATION DEPENDENT MRNA DECAY | 48 | 29 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF RNA | 330 | 155 | All SZGR 2.0 genes in this pathway |
| REACTOME INTERFERON SIGNALING | 159 | 116 | All SZGR 2.0 genes in this pathway |
| REACTOME PKB MEDIATED EVENTS | 29 | 23 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY INSULIN RECEPTOR | 108 | 72 | All SZGR 2.0 genes in this pathway |
| REACTOME IMMUNE SYSTEM | 933 | 616 | All SZGR 2.0 genes in this pathway |
| REACTOME MTORC1 MEDIATED SIGNALLING | 11 | 8 | All SZGR 2.0 genes in this pathway |
| REACTOME CYTOKINE SIGNALING IN IMMUNE SYSTEM | 270 | 204 | All SZGR 2.0 genes in this pathway |
| REACTOME PI3K CASCADE | 71 | 51 | All SZGR 2.0 genes in this pathway |
| PICCALUGA ANGIOIMMUNOBLASTIC LYMPHOMA DN | 136 | 86 | All SZGR 2.0 genes in this pathway |
| ZHOU INFLAMMATORY RESPONSE LIVE UP | 485 | 293 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP | 1382 | 904 | All SZGR 2.0 genes in this pathway |
| TIEN INTESTINE PROBIOTICS 24HR UP | 557 | 331 | All SZGR 2.0 genes in this pathway |
| ZHOU INFLAMMATORY RESPONSE FIMA UP | 544 | 308 | All SZGR 2.0 genes in this pathway |
| ZHOU INFLAMMATORY RESPONSE LPS UP | 431 | 237 | All SZGR 2.0 genes in this pathway |
| BIDUS METASTASIS UP | 214 | 134 | All SZGR 2.0 genes in this pathway |
| GOZGIT ESR1 TARGETS DN | 781 | 465 | All SZGR 2.0 genes in this pathway |
| CHOW RASSF1 TARGETS UP | 27 | 17 | All SZGR 2.0 genes in this pathway |
| CAIRO PML TARGETS BOUND BY MYC UP | 23 | 17 | All SZGR 2.0 genes in this pathway |
| SCHLOSSER MYC TARGETS REPRESSED BY SERUM | 159 | 93 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 DN | 855 | 609 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 COMMON DN | 483 | 336 | All SZGR 2.0 genes in this pathway |
| LIANG HEMATOPOIESIS STEM CELL NUMBER SMALL VS HUGE UP | 38 | 29 | All SZGR 2.0 genes in this pathway |
| PUJANA CHEK2 PCC NETWORK | 779 | 480 | All SZGR 2.0 genes in this pathway |
| BENPORATH MYC TARGETS WITH EBOX | 230 | 156 | All SZGR 2.0 genes in this pathway |
| BENPORATH CYCLING GENES | 648 | 385 | All SZGR 2.0 genes in this pathway |
| STARK PREFRONTAL CORTEX 22Q11 DELETION DN | 517 | 309 | All SZGR 2.0 genes in this pathway |
| MORI SMALL PRE BII LYMPHOCYTE DN | 76 | 52 | All SZGR 2.0 genes in this pathway |
| SHEPARD CRUSH AND BURN MUTANT UP | 197 | 110 | All SZGR 2.0 genes in this pathway |
| SHEPARD BMYB MORPHOLINO UP | 205 | 126 | All SZGR 2.0 genes in this pathway |
| BROCKE APOPTOSIS REVERSED BY IL6 | 144 | 98 | All SZGR 2.0 genes in this pathway |
| FERNANDEZ BOUND BY MYC | 182 | 116 | All SZGR 2.0 genes in this pathway |
| BROWN MYELOID CELL DEVELOPMENT DN | 129 | 86 | All SZGR 2.0 genes in this pathway |
| XU RESPONSE TO TRETINOIN AND NSC682994 DN | 15 | 12 | All SZGR 2.0 genes in this pathway |
| DEBIASI APOPTOSIS BY REOVIRUS INFECTION UP | 314 | 201 | All SZGR 2.0 genes in this pathway |
| WANG SMARCE1 TARGETS DN | 371 | 218 | All SZGR 2.0 genes in this pathway |
| ZHU CMV ALL UP | 120 | 89 | All SZGR 2.0 genes in this pathway |
| GENTILE UV RESPONSE CLUSTER D2 | 41 | 30 | All SZGR 2.0 genes in this pathway |
| YIH RESPONSE TO ARSENITE C4 | 18 | 12 | All SZGR 2.0 genes in this pathway |
| GENTILE UV HIGH DOSE DN | 312 | 203 | All SZGR 2.0 genes in this pathway |
| ZHU CMV 24 HR UP | 93 | 65 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 6HR DN | 911 | 527 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 24HR DN | 1011 | 592 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 STIMULATED | 1022 | 619 | All SZGR 2.0 genes in this pathway |
| KYNG DNA DAMAGE DN | 195 | 135 | All SZGR 2.0 genes in this pathway |
| KYNG ENVIRONMENTAL STRESS RESPONSE UP | 56 | 41 | All SZGR 2.0 genes in this pathway |
| DAIRKEE CANCER PRONE RESPONSE BPA E2 | 118 | 65 | All SZGR 2.0 genes in this pathway |
| ZHANG TLX TARGETS 60HR DN | 277 | 166 | All SZGR 2.0 genes in this pathway |
| DANG MYC TARGETS UP | 143 | 100 | All SZGR 2.0 genes in this pathway |
| DANG BOUND BY MYC | 1103 | 714 | All SZGR 2.0 genes in this pathway |
| WHITFIELD CELL CYCLE M G1 | 148 | 95 | All SZGR 2.0 genes in this pathway |
| THILLAINADESAN ZNF217 TARGETS UP | 44 | 22 | All SZGR 2.0 genes in this pathway |
| JOHNSTONE PARVB TARGETS 3 DN | 918 | 550 | All SZGR 2.0 genes in this pathway |
| LEE BMP2 TARGETS DN | 882 | 538 | All SZGR 2.0 genes in this pathway |
| KOINUMA TARGETS OF SMAD2 OR SMAD3 | 824 | 528 | All SZGR 2.0 genes in this pathway |
| IKEDA MIR1 TARGETS DN | 7 | 7 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-1/206 | 790 | 796 | m8 | hsa-miR-1 | UGGAAUGUAAAGAAGUAUGUA |
| hsa-miR-206SZ | UGGAAUGUAAGGAAGUGUGUGG | ||||
| hsa-miR-613 | AGGAAUGUUCCUUCUUUGCC | ||||
| miR-15/16/195/424/497 | 496 | 502 | 1A | hsa-miR-15abrain | UAGCAGCACAUAAUGGUUUGUG |
| hsa-miR-16brain | UAGCAGCACGUAAAUAUUGGCG | ||||
| hsa-miR-15bbrain | UAGCAGCACAUCAUGGUUUACA | ||||
| hsa-miR-195SZ | UAGCAGCACAGAAAUAUUGGC | ||||
| hsa-miR-424 | CAGCAGCAAUUCAUGUUUUGAA | ||||
| hsa-miR-497 | CAGCAGCACACUGUGGUUUGU | ||||
| miR-150 | 52 | 58 | m8 | hsa-miR-150 | UCUCCCAACCCUUGUACCAGUG |
| miR-186 | 652 | 658 | m8 | hsa-miR-186 | CAAAGAAUUCUCCUUUUGGGCUU |
| miR-203.1 | 709 | 715 | 1A | hsa-miR-203 | UGAAAUGUUUAGGACCACUAG |
| miR-205 | 601 | 607 | 1A | hsa-miR-205 | UCCUUCAUUCCACCGGAGUCUG |
| miR-330 | 869 | 875 | m8 | hsa-miR-330brain | GCAAAGCACACGGCCUGCAGAGA |
| miR-376c | 729 | 735 | m8 | hsa-miR-376c | AACAUAGAGGAAAUUCCACG |
| miR-377 | 737 | 743 | m8 | hsa-miR-377 | AUCACACAAAGGCAACUUUUGU |
| miR-493-5p | 924 | 930 | 1A | hsa-miR-493-5p | UUGUACAUGGUAGGCUUUCAUU |
| miR-503 | 496 | 502 | 1A | hsa-miR-503 | UAGCAGCGGGAACAGUUCUGCAG |
| miR-9 | 63 | 69 | m8 | hsa-miR-9SZ | UCUUUGGUUAUCUAGCUGUAUGA |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.