Gene Page: ELAVL4
Summary ?
| GeneID | 1996 |
| Symbol | ELAVL4 |
| Synonyms | HUD|PNEM |
| Description | ELAV like neuron-specific RNA binding protein 4 |
| Reference | MIM:168360|HGNC:HGNC:3315|Ensembl:ENSG00000162374|HPRD:01343|Vega:OTTHUMG00000007877 |
| Gene type | protein-coding |
| Map location | 1p34 |
| Pascal p-value | 4.049E-5 |
| Sherlock p-value | 0.952 |
| Fetal beta | 0.282 |
| eGene | Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs7132043 | chr12 | 80968399 | ELAVL4 | 1996 | 0.08 | trans |
Section II. Transcriptome annotation
General gene expression (GTEx)
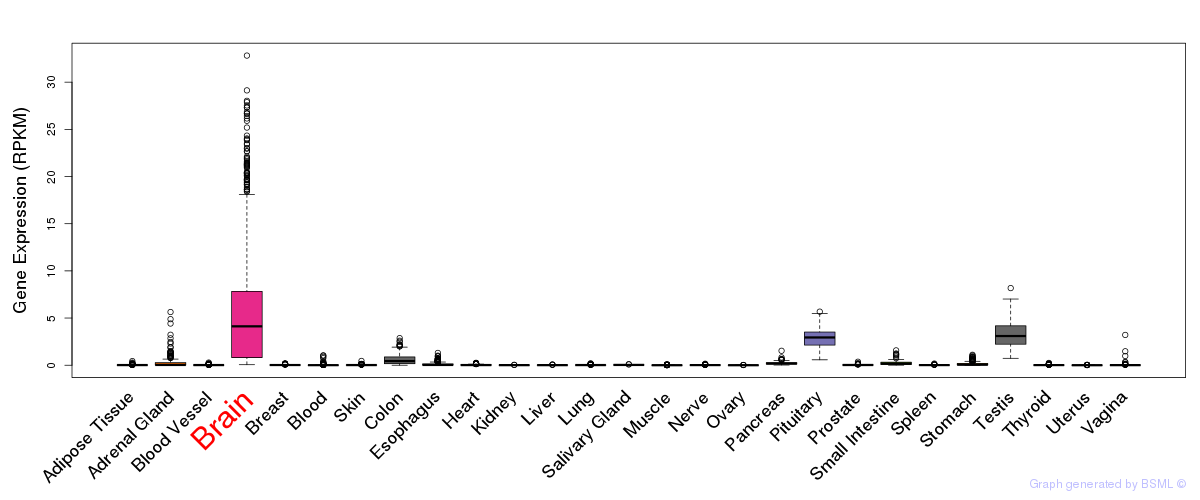
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AL117209.1 | 0.94 | 0.94 |
| UBE2O | 0.92 | 0.92 |
| SIN3B | 0.91 | 0.92 |
| BRPF3 | 0.91 | 0.92 |
| TECPR1 | 0.91 | 0.92 |
| ARFGAP1 | 0.91 | 0.89 |
| AP3D1 | 0.91 | 0.90 |
| DNAJC11 | 0.90 | 0.87 |
| BRSK2 | 0.90 | 0.90 |
| RUNDC1 | 0.90 | 0.90 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.21 | -0.79 | -0.75 |
| AF347015.31 | -0.77 | -0.70 |
| MT-CO2 | -0.76 | -0.69 |
| AF347015.8 | -0.73 | -0.67 |
| C1orf54 | -0.72 | -0.78 |
| HIGD1B | -0.72 | -0.66 |
| MT-CYB | -0.72 | -0.64 |
| AF347015.33 | -0.71 | -0.63 |
| AF347015.27 | -0.71 | -0.67 |
| AF347015.2 | -0.70 | -0.60 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| LIU SOX4 TARGETS UP | 137 | 94 | All SZGR 2.0 genes in this pathway |
| LASTOWSKA NEUROBLASTOMA COPY NUMBER DN | 800 | 473 | All SZGR 2.0 genes in this pathway |
| MARTORIATI MDM4 TARGETS NEUROEPITHELIUM DN | 164 | 111 | All SZGR 2.0 genes in this pathway |
| SHETH LIVER CANCER VS TXNIP LOSS PAM5 | 94 | 59 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 12HR DN | 57 | 45 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER SOX9 TARGETS IN PROSTATE DEVELOPMENT DN | 45 | 33 | All SZGR 2.0 genes in this pathway |
| SHEPARD BMYB MORPHOLINO DN | 200 | 112 | All SZGR 2.0 genes in this pathway |
| SHEPARD BMYB TARGETS | 74 | 41 | All SZGR 2.0 genes in this pathway |
| SHEPARD CRUSH AND BURN MUTANT DN | 185 | 111 | All SZGR 2.0 genes in this pathway |
| POMEROY MEDULLOBLASTOMA DESMOPLASIC VS CLASSIC DN | 59 | 35 | All SZGR 2.0 genes in this pathway |
| MAGRANGEAS MULTIPLE MYELOMA IGLL VS IGLK UP | 42 | 24 | All SZGR 2.0 genes in this pathway |
| ABE VEGFA TARGETS 2HR | 34 | 21 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS STEM CELL LONG TERM | 302 | 191 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE DN | 1237 | 837 | All SZGR 2.0 genes in this pathway |
| XU GH1 EXOGENOUS TARGETS DN | 120 | 69 | All SZGR 2.0 genes in this pathway |
| ABE VEGFA TARGETS 30MIN | 29 | 21 | All SZGR 2.0 genes in this pathway |
| LABBE TGFB1 TARGETS UP | 102 | 64 | All SZGR 2.0 genes in this pathway |
| LIN NPAS4 TARGETS DN | 68 | 48 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS UP | 1305 | 895 | All SZGR 2.0 genes in this pathway |
| GYORFFY MITOXANTRONE RESISTANCE | 57 | 35 | All SZGR 2.0 genes in this pathway |
| DANG REGULATED BY MYC DN | 253 | 192 | All SZGR 2.0 genes in this pathway |