Gene Page: EMD
Summary ?
| GeneID | 2010 |
| Symbol | EMD |
| Synonyms | EDMD|LEMD5|STA |
| Description | emerin |
| Reference | MIM:300384|HGNC:HGNC:3331|Ensembl:ENSG00000102119|HPRD:02309|Vega:OTTHUMG00000033186 |
| Gene type | protein-coding |
| Map location | Xq28 |
| Sherlock p-value | 0.81 |
| Fetal beta | 0.318 |
| DMG | 1 (# studies) |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:vanEijk_2014 | Genome-wide DNA methylation analysis | This dataset includes 432 differentially methylated CpG sites corresponding to 391 unique transcripts between schizophrenia patients (n=260) and unaffected controls (n=250). | 1 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenics,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg09229960 | X | 153607856 | EMD | 1.712E-4 | 5.115 | DMG:vanEijk_2014 |
Section II. Transcriptome annotation
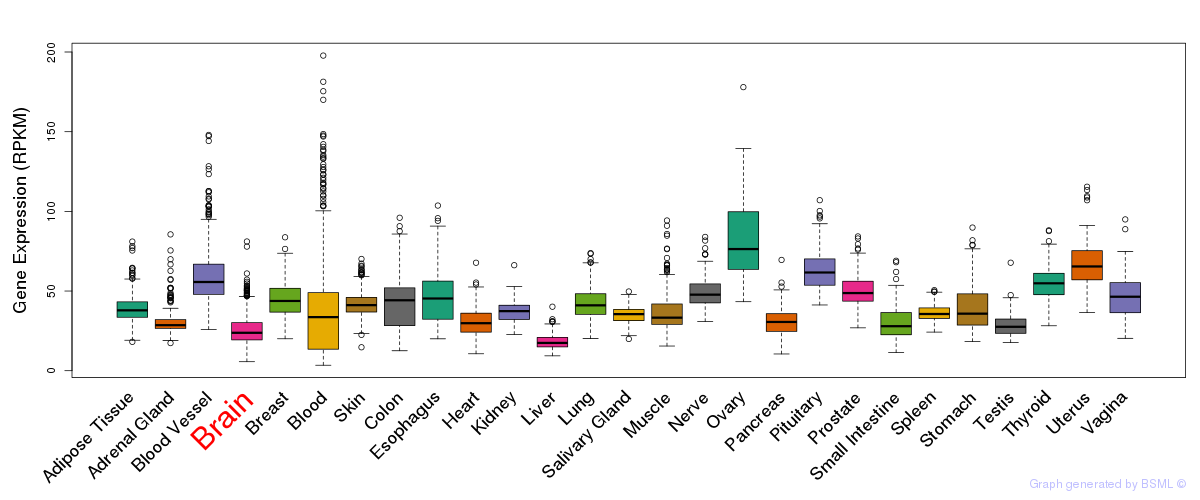
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ACTB | PS1TP5BP1 | actin, beta | - | HPRD | 12670476 |
| ACTG2 | ACT | ACTA3 | ACTE | ACTL3 | ACTSG | actin, gamma 2, smooth muscle, enteric | Affinity Capture-Western Reconstituted Complex | BioGRID | 12670476 |
| AKAP8L | DKFZp434L0650 | HA95 | HAP95 | NAKAP | NAKAP95 | A kinase (PRKA) anchor protein 8-like | - | HPRD,BioGRID | 11034899 |
| BANF1 | BAF | BCRP1 | D14S1460 | MGC111161 | barrier to autointegration factor 1 | Emerin interacts with BAF. | BIND | 11792821 |
| BANF1 | BAF | BCRP1 | D14S1460 | MGC111161 | barrier to autointegration factor 1 | - | HPRD,BioGRID | 11792822 |
| BCLAF1 | BTF | KIAA0164 | bK211L9.1 | BCL2-associated transcription factor 1 | - | HPRD | 15009215 |
| BEND7 | C10orf30 | FLJ40283 | MGC35247 | BEN domain containing 7 | Two-hybrid | BioGRID | 16189514 |
| COPS6 | CSN6 | MOV34-34KD | COP9 constitutive photomorphogenic homolog subunit 6 (Arabidopsis) | Two-hybrid | BioGRID | 16169070 |
| CREB3 | LUMAN | LZIP | MGC15333 | MGC19782 | cAMP responsive element binding protein 3 | Two-hybrid | BioGRID | 16189514 |
| DPPA4 | 2410091M23Rik | FLJ10713 | developmental pluripotency associated 4 | Two-hybrid | BioGRID | 16169070 |
| FATE1 | FATE | fetal and adult testis expressed 1 | Two-hybrid | BioGRID | 16189514 |
| GMCL1 | BTBD13 | GCL | GCL1 | germ cell-less homolog 1 (Drosophila) | - | HPRD,BioGRID | 12493765 |
| LMNA | CDCD1 | CDDC | CMD1A | CMT2B1 | EMD2 | FPL | FPLD | HGPS | IDC | LDP1 | LFP | LGMD1B | LMN1 | LMNC | PRO1 | lamin A/C | - | HPRD,BioGRID | 10673356 |11173535 |
| LMNB1 | ADLD | LMN | LMN2 | LMNB | MGC111419 | lamin B1 | Affinity Capture-Western | BioGRID | 12670476 |
| LPL | HDLCQ11 | LIPD | lipoprotein lipase | Two-hybrid | BioGRID | 12755701 |
| PSME1 | IFI5111 | MGC8628 | PA28A | PA28alpha | REGalpha | proteasome (prosome, macropain) activator subunit 1 (PA28 alpha) | Affinity Capture-Western Two-hybrid | BioGRID | 12755701 |
| SH3GL2 | CNSA2 | EEN-B1 | FLJ20276 | FLJ25015 | SH3D2A | SH3P4 | SH3-domain GRB2-like 2 | Two-hybrid | BioGRID | 16169070 |
| SH3GL3 | CNSA3 | EEN-2B-L3 | EEN-B2 | HsT19371 | SH3D2C | SH3P13 | SH3-domain GRB2-like 3 | Two-hybrid | BioGRID | 16169070 |
| SYNE1 | 8B | CPG2 | DKFZp781J13156 | FLJ30878 | FLJ41140 | KIAA0796 | KIAA1262 | KIAA1756 | MYNE1 | SCAR8 | spectrin repeat containing, nuclear envelope 1 | - | HPRD,BioGRID | 12163176 |
| VAV1 | VAV | vav 1 guanine nucleotide exchange factor | Two-hybrid | BioGRID | 12755701 |
| YTHDC1 | KIAA1966 | YT521 | YT521-B | YTH domain containing 1 | - | HPRD,BioGRID | 12755701 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG HYPERTROPHIC CARDIOMYOPATHY HCM | 85 | 65 | All SZGR 2.0 genes in this pathway |
| KEGG ARRHYTHMOGENIC RIGHT VENTRICULAR CARDIOMYOPATHY ARVC | 76 | 59 | All SZGR 2.0 genes in this pathway |
| KEGG DILATED CARDIOMYOPATHY | 92 | 68 | All SZGR 2.0 genes in this pathway |
| OSMAN BLADDER CANCER DN | 406 | 230 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE KERATINOCYTE UP | 530 | 342 | All SZGR 2.0 genes in this pathway |
| DAIRKEE TERT TARGETS UP | 380 | 213 | All SZGR 2.0 genes in this pathway |
| SPIELMAN LYMPHOBLAST EUROPEAN VS ASIAN UP | 479 | 299 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 12HR UP | 116 | 79 | All SZGR 2.0 genes in this pathway |
| PENG LEUCINE DEPRIVATION DN | 187 | 122 | All SZGR 2.0 genes in this pathway |
| BURTON ADIPOGENESIS PEAK AT 2HR | 51 | 36 | All SZGR 2.0 genes in this pathway |
| WEST ADRENOCORTICAL TUMOR DN | 546 | 362 | All SZGR 2.0 genes in this pathway |
| WANG TUMOR INVASIVENESS DN | 210 | 128 | All SZGR 2.0 genes in this pathway |
| COULOUARN TEMPORAL TGFB1 SIGNATURE UP | 109 | 68 | All SZGR 2.0 genes in this pathway |
| HOSHIDA LIVER CANCER SURVIVAL DN | 113 | 76 | All SZGR 2.0 genes in this pathway |
| WIERENGA PML INTERACTOME | 42 | 23 | All SZGR 2.0 genes in this pathway |
| ROESSLER LIVER CANCER METASTASIS UP | 107 | 72 | All SZGR 2.0 genes in this pathway |