Gene Page: RAB12
Summary ?
| GeneID | 201475 |
| Symbol | RAB12 |
| Synonyms | - |
| Description | RAB12, member RAS oncogene family |
| Reference | MIM:616448|HGNC:HGNC:31332|Ensembl:ENSG00000206418|Vega:OTTHUMG00000178949 |
| Gene type | protein-coding |
| Map location | 18p11.22 |
| Pascal p-value | 0.922 |
| Fetal beta | 1.653 |
| eGene | Myers' cis & trans |
| Support | INTRACELLULAR TRAFFICKING G2Cdb.humanPSD G2Cdb.humanPSP |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs6710928 | chr2 | 4962490 | RAB12 | 201475 | 0.03 | trans | ||
| rs5009573 | chr2 | 4963241 | RAB12 | 201475 | 0.01 | trans | ||
| rs13401024 | chr2 | 158849591 | RAB12 | 201475 | 0.09 | trans | ||
| rs17065046 | chr13 | 76366621 | RAB12 | 201475 | 0.18 | trans | ||
| rs4885350 | chr13 | 76375316 | RAB12 | 201475 | 0.03 | trans | ||
| rs4884020 | chr13 | 76379296 | RAB12 | 201475 | 0.04 | trans | ||
| rs17065075 | chr13 | 76384561 | RAB12 | 201475 | 0.07 | trans | ||
| rs17065132 | chr13 | 76421681 | RAB12 | 201475 | 0.07 | trans | ||
| rs2289063 | chr16 | 75667790 | RAB12 | 201475 | 0.2 | trans | ||
| rs4629073 | chr18 | 67487570 | RAB12 | 201475 | 0.14 | trans | ||
| rs6030274 | chr20 | 41118094 | RAB12 | 201475 | 0.01 | trans | ||
| rs6016807 | chr20 | 41118122 | RAB12 | 201475 | 0.01 | trans | ||
| rs6093659 | chr20 | 41121506 | RAB12 | 201475 | 0.01 | trans | ||
| rs6093660 | chr20 | 41121608 | RAB12 | 201475 | 0.01 | trans | ||
| rs6016811 | chr20 | 41127168 | RAB12 | 201475 | 0.01 | trans | ||
| rs10521726 | chrX | 123667220 | RAB12 | 201475 | 0.08 | trans |
Section II. Transcriptome annotation
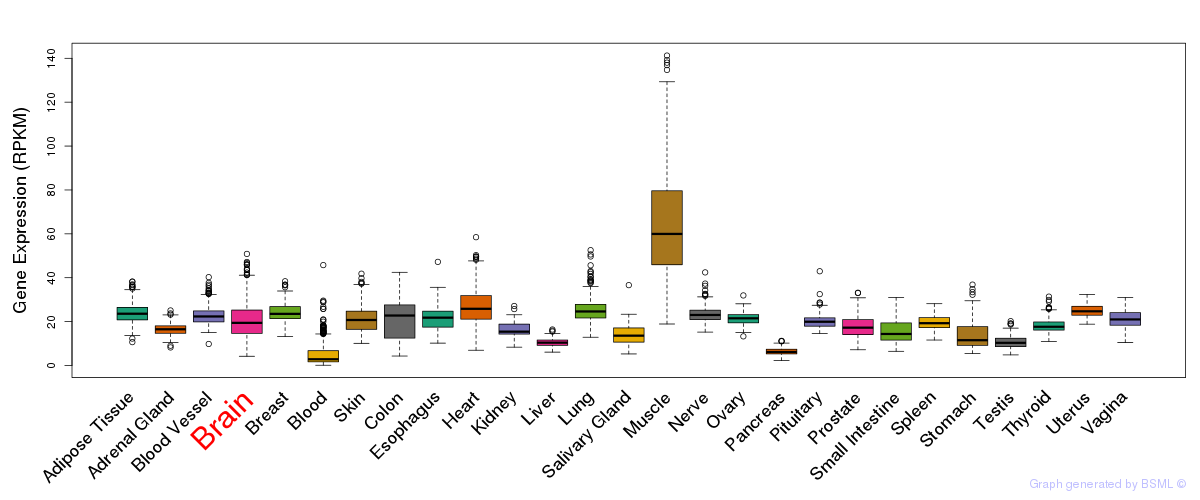
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| TAKEDA TARGETS OF NUP98 HOXA9 FUSION 6HR DN | 41 | 29 | All SZGR 2.0 genes in this pathway |
| FALVELLA SMOKERS WITH LUNG CANCER | 80 | 52 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 6HR DN | 514 | 330 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY E2F4 UNSTIMULATED | 728 | 415 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 TARGETS UP | 673 | 430 | All SZGR 2.0 genes in this pathway |
| MARTINEZ TP53 TARGETS DN | 593 | 372 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 AND TP53 TARGETS DN | 591 | 366 | All SZGR 2.0 genes in this pathway |
| IZADPANAH STEM CELL ADIPOSE VS BONE DN | 108 | 68 | All SZGR 2.0 genes in this pathway |
| SWEET LUNG CANCER KRAS DN | 435 | 289 | All SZGR 2.0 genes in this pathway |
| CHANDRAN METASTASIS UP | 221 | 135 | All SZGR 2.0 genes in this pathway |
| JOHNSTONE PARVB TARGETS 3 DN | 918 | 550 | All SZGR 2.0 genes in this pathway |
| RATTENBACHER BOUND BY CELF1 | 467 | 251 | All SZGR 2.0 genes in this pathway |