Gene Page: CTTN
Summary ?
| GeneID | 2017 |
| Symbol | CTTN |
| Synonyms | EMS1 |
| Description | cortactin |
| Reference | MIM:164765|HGNC:HGNC:3338|Ensembl:ENSG00000085733|HPRD:01268|Vega:OTTHUMG00000134307 |
| Gene type | protein-coding |
| Map location | 11q13 |
| Pascal p-value | 0.195 |
| Sherlock p-value | 0.823 |
| Fetal beta | -0.259 |
| DMG | 1 (# studies) |
| eGene | Myers' cis & trans |
| Support | CELL ADHESION AND TRANSSYNAPTIC SIGNALING G2Cdb.humanNRC G2Cdb.humanPSD G2Cdb.humanPSP Ascano FMRP targets Potential synaptic genes |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Nishioka_2013 | Genome-wide DNA methylation analysis | The authors investigated the methylation profiles of DNA in peripheral blood cells from 18 patients with first-episode schizophrenia (FESZ) and from 15 normal controls. | 1 |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.0836 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg15383392 | 11 | 70244691 | CTTN | -0.022 | 0.55 | DMG:Nishioka_2013 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs7120675 | chr11 | 71050680 | CTTN | 2017 | 0.2 | cis |
Section II. Transcriptome annotation
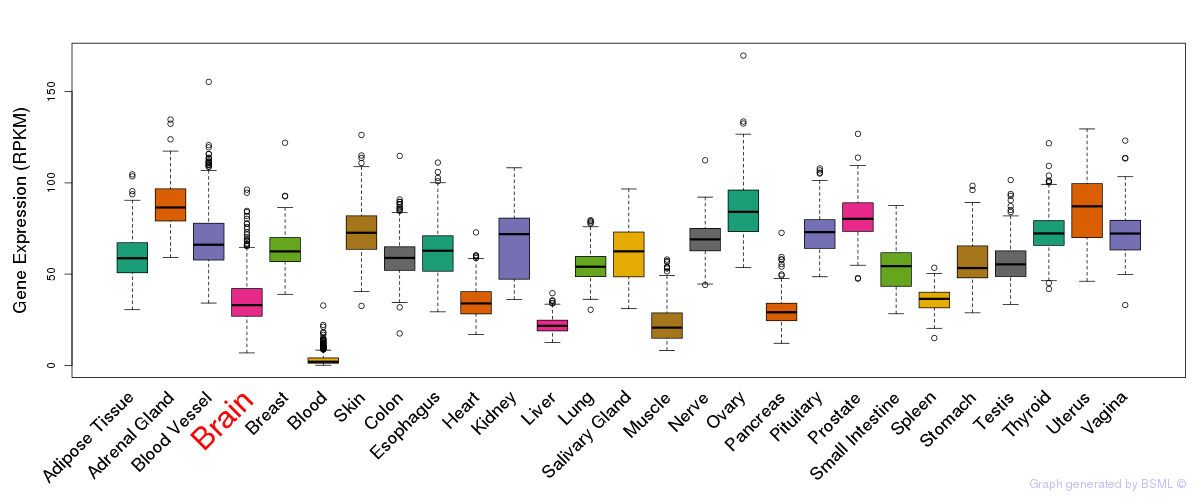
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005515 | protein binding | IEA | - | |
| GO:0005515 | protein binding | ISS | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0001726 | ruffle | ISS | - | |
| GO:0005856 | cytoskeleton | TAS | 9054437 | |
| GO:0005625 | soluble fraction | TAS | 9054437 | |
| GO:0005737 | cytoplasm | IEA | - | |
| GO:0005737 | cytoplasm | ISS | - | |
| GO:0030027 | lamellipodium | IEA | - | |
| GO:0030027 | lamellipodium | ISS | - | |
| GO:0005938 | cell cortex | IEA | - | |
| GO:0005938 | cell cortex | ISS | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ACTR2 | ARP2 | ARP2 actin-related protein 2 homolog (yeast) | Affinity Capture-MS | BioGRID | 11018051 |
| ACTR3 | ARP3 | ARP3 actin-related protein 3 homolog (yeast) | - | HPRD,BioGRID | 12176742 |
| ARHGAP17 | DKFZp564A1363 | FLJ37567 | FLJ43368 | MGC87805 | MST066 | MST110 | MSTP038 | MSTP066 | MSTP110 | NADRIN | RICH1 | WBP15 | Rho GTPase activating protein 17 | Reconstituted Complex | BioGRID | 11431473 |
| ARHGAP8 | BPGAP1 | FLJ20185 | PP610 | Rho GTPase activating protein 8 | BPGAP1 interacts with Cortactin. | BIND | 15064355 |
| ARPC2 | ARC34 | PNAS-139 | PRO2446 | p34-Arc | actin related protein 2/3 complex, subunit 2, 34kDa | Affinity Capture-MS Affinity Capture-Western | BioGRID | 11018051 |
| ARPC3 | ARC21 | p21-Arc | actin related protein 2/3 complex, subunit 3, 21kDa | Affinity Capture-MS | BioGRID | 11018051 |
| ARPC4 | ARC20 | MGC13544 | p20-Arc | actin related protein 2/3 complex, subunit 4, 20kDa | Affinity Capture-MS | BioGRID | 11018051 |
| ASAP1 | AMAP1 | CENTB4 | DDEF1 | KIAA1249 | PAG2 | PAP | ZG14P | ArfGAP with SH3 domain, ankyrin repeat and PH domain 1 | DDEF1 (AMAP1) interacts with an unspecified isoform of CTTN (cortactin). | BIND | 15719014 |
| CASP3 | CPP32 | CPP32B | SCA-1 | caspase 3, apoptosis-related cysteine peptidase | Biochemical Activity | BioGRID | 11689006 |
| CHD3 | Mi-2a | Mi2-ALPHA | ZFH | chromodomain helicase DNA binding protein 3 | Two-hybrid | BioGRID | 16169070 |
| CRB2 | FLJ16786 | FLJ38464 | crumbs homolog 2 (Drosophila) | Affinity Capture-Western | BioGRID | 11439336 |
| CTNND1 | CAS | CTNND | KIAA0384 | P120CAS | P120CTN | p120 | catenin (cadherin-associated protein), delta 1 | - | HPRD,BioGRID | 12835311 |
| CTNND2 | GT24 | NPRAP | catenin (cadherin-associated protein), delta 2 (neural plakophilin-related arm-repeat protein) | - | HPRD | 12835311 |
| DNM1 | DNM | dynamin 1 | - | HPRD | 12419186 |
| DNM2 | CMTDI1 | CMTDIB | DI-CMTB | DYN2 | DYNII | dynamin 2 | - | HPRD | 12419186 |
| FER | TYK3 | fer (fps/fes related) tyrosine kinase | - | HPRD,BioGRID | 9722593 |
| FGD1 | AAS | FGDY | ZFYVE3 | FYVE, RhoGEF and PH domain containing 1 | - | HPRD | 12913069 |
| GRB2 | ASH | EGFRBP-GRB2 | Grb3-3 | MST084 | MSTP084 | growth factor receptor-bound protein 2 | - | HPRD | 11439336 |
| KCNA2 | HBK5 | HK4 | HUKIV | KV1.2 | MGC50217 | MK2 | NGK1 | RBK2 | potassium voltage-gated channel, shaker-related subfamily, member 2 | - | HPRD,BioGRID | 12151401 |
| MYLK | DKFZp686I10125 | FLJ12216 | KRP | MLCK | MLCK1 | MLCK108 | MLCK210 | MSTP083 | MYLK1 | smMLCK | myosin light chain kinase | - | HPRD,BioGRID | 12408982 |
| PXN | FLJ16691 | paxillin | PXN (Paxillin) interacts with an unspecified isoform of CTTN (cortactin). | BIND | 15719014 |
| SDC3 | N-syndecan | SDCN | SYND3 | syndecan 3 | - | HPRD,BioGRID | 9553134 |
| SHANK2 | CORTBP1 | CTTNBP1 | ProSAP1 | SHANK | SPANK-3 | SH3 and multiple ankyrin repeat domains 2 | - | HPRD,BioGRID | 9742101 |
| SYK | DKFZp313N1010 | FLJ25043 | FLJ37489 | spleen tyrosine kinase | - | HPRD,BioGRID | 8636079 |
| TJP1 | DKFZp686M05161 | MGC133289 | ZO-1 | tight junction protein 1 (zona occludens 1) | - | HPRD | 9792678|11018051 |
| WASL | DKFZp779G0847 | MGC48327 | N-WASP | NWASP | Wiskott-Aldrich syndrome-like | - | HPRD,BioGRID | 11830518 |
| WIPF1 | MGC111041 | PRPL-2 | WASPIP | WIP | WAS/WASL interacting protein family, member 1 | - | HPRD,BioGRID | 12620186 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG TIGHT JUNCTION | 134 | 86 | All SZGR 2.0 genes in this pathway |
| KEGG PATHOGENIC ESCHERICHIA COLI INFECTION | 59 | 36 | All SZGR 2.0 genes in this pathway |
| BIOCARTA AGR PATHWAY | 36 | 31 | All SZGR 2.0 genes in this pathway |
| PID ECADHERIN NASCENT AJ PATHWAY | 39 | 33 | All SZGR 2.0 genes in this pathway |
| PID PDGFRB PATHWAY | 129 | 103 | All SZGR 2.0 genes in this pathway |
| PID SYNDECAN 3 PATHWAY | 17 | 15 | All SZGR 2.0 genes in this pathway |
| PID NCADHERIN PATHWAY | 33 | 32 | All SZGR 2.0 genes in this pathway |
| PID FGF PATHWAY | 55 | 37 | All SZGR 2.0 genes in this pathway |
| PYEON HPV POSITIVE TUMORS DN | 10 | 5 | All SZGR 2.0 genes in this pathway |
| ONKEN UVEAL MELANOMA UP | 783 | 507 | All SZGR 2.0 genes in this pathway |
| BORCZUK MALIGNANT MESOTHELIOMA DN | 104 | 59 | All SZGR 2.0 genes in this pathway |
| GINESTIER BREAST CANCER 20Q13 AMPLIFICATION UP | 119 | 66 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC1 TARGETS UP | 457 | 269 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC3 TARGETS UP | 501 | 327 | All SZGR 2.0 genes in this pathway |
| NAGASHIMA NRG1 SIGNALING UP | 176 | 123 | All SZGR 2.0 genes in this pathway |
| JAATINEN HEMATOPOIETIC STEM CELL DN | 226 | 132 | All SZGR 2.0 genes in this pathway |
| DELYS THYROID CANCER UP | 443 | 294 | All SZGR 2.0 genes in this pathway |
| CHIN BREAST CANCER COPY NUMBER UP | 27 | 18 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN DN | 1781 | 1082 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN RESPONSE TO MC AND DOXORUBICIN DN | 770 | 415 | All SZGR 2.0 genes in this pathway |
| MAHADEVAN RESPONSE TO MP470 DN | 19 | 11 | All SZGR 2.0 genes in this pathway |
| SNIJDERS AMPLIFIED IN HEAD AND NECK TUMORS | 37 | 27 | All SZGR 2.0 genes in this pathway |
| MARTORIATI MDM4 TARGETS FETAL LIVER DN | 514 | 319 | All SZGR 2.0 genes in this pathway |
| MURATA VIRULENCE OF H PILORI | 24 | 16 | All SZGR 2.0 genes in this pathway |
| SHETH LIVER CANCER VS TXNIP LOSS PAM1 | 229 | 137 | All SZGR 2.0 genes in this pathway |
| SPIELMAN LYMPHOBLAST EUROPEAN VS ASIAN UP | 479 | 299 | All SZGR 2.0 genes in this pathway |
| NUYTTEN EZH2 TARGETS UP | 1037 | 673 | All SZGR 2.0 genes in this pathway |
| NAKAYAMA FRA2 TARGETS | 43 | 27 | All SZGR 2.0 genes in this pathway |
| LOPEZ MBD TARGETS | 957 | 597 | All SZGR 2.0 genes in this pathway |
| LIU NASOPHARYNGEAL CARCINOMA | 70 | 38 | All SZGR 2.0 genes in this pathway |
| WEI MYCN TARGETS WITH E BOX | 795 | 478 | All SZGR 2.0 genes in this pathway |
| NIKOLSKY BREAST CANCER 11Q12 Q14 AMPLICON | 158 | 93 | All SZGR 2.0 genes in this pathway |
| DING LUNG CANCER EXPRESSION BY COPY NUMBER | 100 | 62 | All SZGR 2.0 genes in this pathway |
| ONDER CDH1 TARGETS 1 UP | 140 | 85 | All SZGR 2.0 genes in this pathway |
| PENG GLUCOSE DEPRIVATION UP | 48 | 26 | All SZGR 2.0 genes in this pathway |
| PENG RAPAMYCIN RESPONSE UP | 203 | 130 | All SZGR 2.0 genes in this pathway |
| STONER ESOPHAGEAL CARCINOGENESIS UP | 39 | 25 | All SZGR 2.0 genes in this pathway |
| ZHAN MULTIPLE MYELOMA LB UP | 48 | 28 | All SZGR 2.0 genes in this pathway |
| MENSE HYPOXIA UP | 98 | 71 | All SZGR 2.0 genes in this pathway |
| REN ALVEOLAR RHABDOMYOSARCOMA DN | 408 | 274 | All SZGR 2.0 genes in this pathway |
| KAAB FAILED HEART ATRIUM DN | 141 | 99 | All SZGR 2.0 genes in this pathway |
| RAMALHO STEMNESS UP | 206 | 118 | All SZGR 2.0 genes in this pathway |
| SIMBULAN PARP1 TARGETS UP | 31 | 23 | All SZGR 2.0 genes in this pathway |
| WEIGEL OXIDATIVE STRESS BY HNE AND TBH | 60 | 42 | All SZGR 2.0 genes in this pathway |
| BURTON ADIPOGENESIS 7 | 51 | 34 | All SZGR 2.0 genes in this pathway |
| RAMPON ENRICHED LEARNING ENVIRONMENT LATE UP | 22 | 16 | All SZGR 2.0 genes in this pathway |
| JI RESPONSE TO FSH UP | 74 | 56 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 6HR UP | 953 | 554 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 24HR UP | 783 | 442 | All SZGR 2.0 genes in this pathway |
| GAVIN FOXP3 TARGETS CLUSTER P4 | 100 | 62 | All SZGR 2.0 genes in this pathway |
| FOSTER TOLERANT MACROPHAGE UP | 156 | 92 | All SZGR 2.0 genes in this pathway |
| HELLER HDAC TARGETS UP | 317 | 208 | All SZGR 2.0 genes in this pathway |
| AMUNDSON POOR SURVIVAL AFTER GAMMA RADIATION 2G | 171 | 96 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 TARGETS DN | 543 | 317 | All SZGR 2.0 genes in this pathway |
| MARTINEZ TP53 TARGETS UP | 602 | 364 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 AND TP53 TARGETS UP | 601 | 369 | All SZGR 2.0 genes in this pathway |
| SAGIV CD24 TARGETS DN | 46 | 26 | All SZGR 2.0 genes in this pathway |
| GRADE COLON CANCER UP | 871 | 505 | All SZGR 2.0 genes in this pathway |
| WEST ADRENOCORTICAL TUMOR DN | 546 | 362 | All SZGR 2.0 genes in this pathway |
| CLIMENT BREAST CANCER COPY NUMBER UP | 23 | 20 | All SZGR 2.0 genes in this pathway |
| BOYLAN MULTIPLE MYELOMA C D DN | 252 | 155 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS DN | 366 | 257 | All SZGR 2.0 genes in this pathway |
| SHEDDEN LUNG CANCER GOOD SURVIVAL A12 | 317 | 177 | All SZGR 2.0 genes in this pathway |
| FONTAINE PAPILLARY THYROID CARCINOMA UP | 66 | 38 | All SZGR 2.0 genes in this pathway |
| RAGHAVACHARI PLATELET SPECIFIC GENES | 70 | 46 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS UP | 504 | 321 | All SZGR 2.0 genes in this pathway |
| JOHNSTONE PARVB TARGETS 3 UP | 430 | 288 | All SZGR 2.0 genes in this pathway |
| JOHNSTONE PARVB TARGETS 3 DN | 918 | 550 | All SZGR 2.0 genes in this pathway |
| FEVR CTNNB1 TARGETS UP | 682 | 433 | All SZGR 2.0 genes in this pathway |
| TORCHIA TARGETS OF EWSR1 FLI1 FUSION UP | 271 | 165 | All SZGR 2.0 genes in this pathway |
| PURBEY TARGETS OF CTBP1 NOT SATB1 DN | 448 | 282 | All SZGR 2.0 genes in this pathway |
| PEDERSEN METASTASIS BY ERBB2 ISOFORM 7 | 403 | 240 | All SZGR 2.0 genes in this pathway |
| DELACROIX RARG BOUND MEF | 367 | 231 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-221/222 | 1348 | 1354 | 1A | hsa-miR-221brain | AGCUACAUUGUCUGCUGGGUUUC |
| hsa-miR-222brain | AGCUACAUCUGGCUACUGGGUCUC | ||||
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.