Gene Page: EPHB4
Summary ?
| GeneID | 2050 |
| Symbol | EPHB4 |
| Synonyms | HTK|MYK1|TYRO11 |
| Description | EPH receptor B4 |
| Reference | MIM:600011|HGNC:HGNC:3395|Ensembl:ENSG00000196411|HPRD:02481|Vega:OTTHUMG00000157040 |
| Gene type | protein-coding |
| Map location | 7q22 |
| Pascal p-value | 0.039 |
| Fetal beta | 0.948 |
| DMG | 1 (# studies) |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 1 |
| Expression | Meta-analysis of gene expression | P value: 2.016 | |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 6 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg01869186 | 7 | 100423987 | EPHB4 | 2.273E-4 | -0.283 | 0.036 | DMG:Wockner_2014 |
Section II. Transcriptome annotation
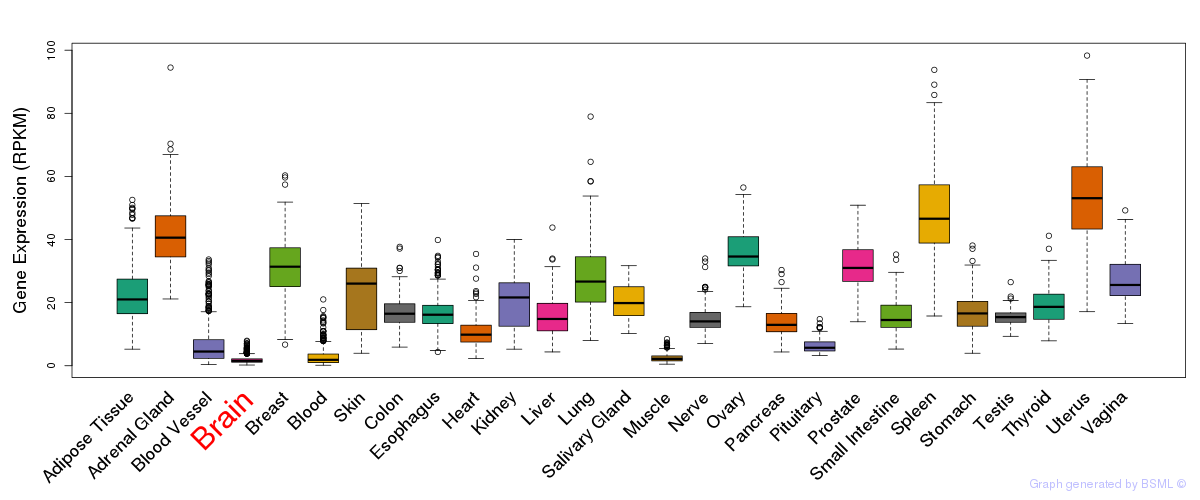
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| TTYH2 | 0.96 | 0.87 |
| C11orf9 | 0.96 | 0.92 |
| LDB3 | 0.95 | 0.91 |
| PLEKHH1 | 0.95 | 0.80 |
| SGK2 | 0.95 | 0.87 |
| GALNT6 | 0.95 | 0.89 |
| SH3TC2 | 0.94 | 0.91 |
| AC022100.1 | 0.94 | 0.87 |
| ATPGD1 | 0.94 | 0.92 |
| C10orf90 | 0.94 | 0.93 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| NR2C2AP | -0.54 | -0.74 |
| MED19 | -0.54 | -0.71 |
| ALKBH2 | -0.54 | -0.74 |
| POLB | -0.53 | -0.74 |
| TRNAU1AP | -0.53 | -0.68 |
| NKIRAS2 | -0.53 | -0.62 |
| HN1 | -0.52 | -0.68 |
| CCDC28B | -0.52 | -0.77 |
| TUBB2B | -0.52 | -0.69 |
| STMN1 | -0.52 | -0.68 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0008046 | axon guidance receptor activity | IEA | axon (GO term level: 6) | - |
| GO:0005102 | receptor binding | IEA | Neurotransmitter (GO term level: 4) | - |
| GO:0000166 | nucleotide binding | IEA | - | |
| GO:0004872 | receptor activity | IEA | - | |
| GO:0005003 | ephrin receptor activity | IEA | - | |
| GO:0005524 | ATP binding | IEA | - | |
| GO:0016740 | transferase activity | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007411 | axon guidance | IEA | axon (GO term level: 13) | - |
| GO:0048170 | positive regulation of long-term neuronal synaptic plasticity | IEA | neuron, neurogenesis, Synap (GO term level: 11) | - |
| GO:0048168 | regulation of neuronal synaptic plasticity | IEA | neuron, Synap (GO term level: 9) | - |
| GO:0050770 | regulation of axonogenesis | IEA | neuron, axon (GO term level: 13) | - |
| GO:0007169 | transmembrane receptor protein tyrosine kinase signaling pathway | IEA | - | |
| GO:0006468 | protein amino acid phosphorylation | IEA | - | |
| GO:0009887 | organ morphogenesis | TAS | 8831703 | |
| GO:0008283 | cell proliferation | TAS | 8152808 | |
| GO:0007612 | learning | IEA | - | |
| GO:0045765 | regulation of angiogenesis | NAS | 16322467 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0016020 | membrane | IEA | - | |
| GO:0016021 | integral to membrane | IEA | - | |
| GO:0009986 | cell surface | IDA | 12493773 | |
| GO:0005887 | integral to plasma membrane | TAS | 8188704 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG AXON GUIDANCE | 129 | 103 | All SZGR 2.0 genes in this pathway |
| PID EPHB FWD PATHWAY | 40 | 38 | All SZGR 2.0 genes in this pathway |
| PID EPHRINB REV PATHWAY | 30 | 25 | All SZGR 2.0 genes in this pathway |
| GAZDA DIAMOND BLACKFAN ANEMIA ERYTHROID DN | 493 | 298 | All SZGR 2.0 genes in this pathway |
| DAVICIONI MOLECULAR ARMS VS ERMS DN | 182 | 111 | All SZGR 2.0 genes in this pathway |
| DEURIG T CELL PROLYMPHOCYTIC LEUKEMIA UP | 368 | 234 | All SZGR 2.0 genes in this pathway |
| UDAYAKUMAR MED1 TARGETS DN | 240 | 171 | All SZGR 2.0 genes in this pathway |
| TIEN INTESTINE PROBIOTICS 2HR UP | 27 | 19 | All SZGR 2.0 genes in this pathway |
| TIEN INTESTINE PROBIOTICS 24HR DN | 214 | 133 | All SZGR 2.0 genes in this pathway |
| WAMUNYOKOLI OVARIAN CANCER LMP UP | 265 | 158 | All SZGR 2.0 genes in this pathway |
| WAMUNYOKOLI OVARIAN CANCER GRADES 1 2 UP | 137 | 84 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA DN | 1375 | 806 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN DN | 1781 | 1082 | All SZGR 2.0 genes in this pathway |
| LOPEZ MBD TARGETS | 957 | 597 | All SZGR 2.0 genes in this pathway |
| AMIT EGF RESPONSE 120 HELA | 69 | 47 | All SZGR 2.0 genes in this pathway |
| MOREAUX MULTIPLE MYELOMA BY TACI UP | 412 | 249 | All SZGR 2.0 genes in this pathway |
| MOREAUX B LYMPHOCYTE MATURATION BY TACI UP | 92 | 58 | All SZGR 2.0 genes in this pathway |
| YAMAZAKI TCEB3 TARGETS UP | 175 | 116 | All SZGR 2.0 genes in this pathway |
| MAHAJAN RESPONSE TO IL1A DN | 76 | 57 | All SZGR 2.0 genes in this pathway |
| VERRECCHIA DELAYED RESPONSE TO TGFB1 | 39 | 26 | All SZGR 2.0 genes in this pathway |
| BURTON ADIPOGENESIS 4 | 48 | 32 | All SZGR 2.0 genes in this pathway |
| HEDENFALK BREAST CANCER BRCA1 VS BRCA2 | 163 | 113 | All SZGR 2.0 genes in this pathway |
| DEBIASI APOPTOSIS BY REOVIRUS INFECTION DN | 287 | 208 | All SZGR 2.0 genes in this pathway |
| BOQUEST STEM CELL DN | 216 | 143 | All SZGR 2.0 genes in this pathway |
| FIRESTEIN PROLIFERATION | 175 | 125 | All SZGR 2.0 genes in this pathway |
| VART KSHV INFECTION ANGIOGENIC MARKERS DN | 138 | 92 | All SZGR 2.0 genes in this pathway |
| MILI PSEUDOPODIA CHEMOTAXIS DN | 457 | 302 | All SZGR 2.0 genes in this pathway |
| MILI PSEUDOPODIA HAPTOTAXIS DN | 668 | 419 | All SZGR 2.0 genes in this pathway |
| BOYAULT LIVER CANCER SUBCLASS G1 UP | 113 | 70 | All SZGR 2.0 genes in this pathway |
| YAGI AML WITH INV 16 TRANSLOCATION | 422 | 277 | All SZGR 2.0 genes in this pathway |
| WONG ADULT TISSUE STEM MODULE | 721 | 492 | All SZGR 2.0 genes in this pathway |
| VERRECCHIA RESPONSE TO TGFB1 C6 | 5 | 5 | All SZGR 2.0 genes in this pathway |
| NOUSHMEHR GBM SOMATIC MUTATED | 9 | 7 | All SZGR 2.0 genes in this pathway |
| VERHAAK GLIOBLASTOMA CLASSICAL | 162 | 122 | All SZGR 2.0 genes in this pathway |
| FEVR CTNNB1 TARGETS DN | 553 | 343 | All SZGR 2.0 genes in this pathway |