Gene Page: ETV6
Summary ?
| GeneID | 2120 |
| Symbol | ETV6 |
| Synonyms | TEL|TEL/ABL|THC5 |
| Description | ETS variant 6 |
| Reference | MIM:600618|HGNC:HGNC:3495|Ensembl:ENSG00000139083|HPRD:15976|Vega:OTTHUMG00000168538 |
| Gene type | protein-coding |
| Map location | 12p13 |
| Pascal p-value | 0.028 |
| Sherlock p-value | 0.388 |
| Fetal beta | -0.267 |
| DMG | 1 (# studies) |
| eGene | Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Nishioka_2013 | Genome-wide DNA methylation analysis | The authors investigated the methylation profiles of DNA in peripheral blood cells from 18 patients with first-episode schizophrenia (FESZ) and from 15 normal controls. | 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg15062795 | 12 | 11802217 | ETV6 | -0.023 | 0.43 | DMG:Nishioka_2013 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs6893753 | chr5 | 35451871 | ETV6 | 2120 | 0.1 | trans | ||
| rs3812061 | chr5 | 126213763 | ETV6 | 2120 | 0.03 | trans | ||
| rs12655258 | chr5 | 126267350 | ETV6 | 2120 | 0.2 | trans | ||
| rs17165234 | chr5 | 126403634 | ETV6 | 2120 | 0.01 | trans | ||
| rs12655006 | chr5 | 126407601 | ETV6 | 2120 | 0.11 | trans |
Section II. Transcriptome annotation
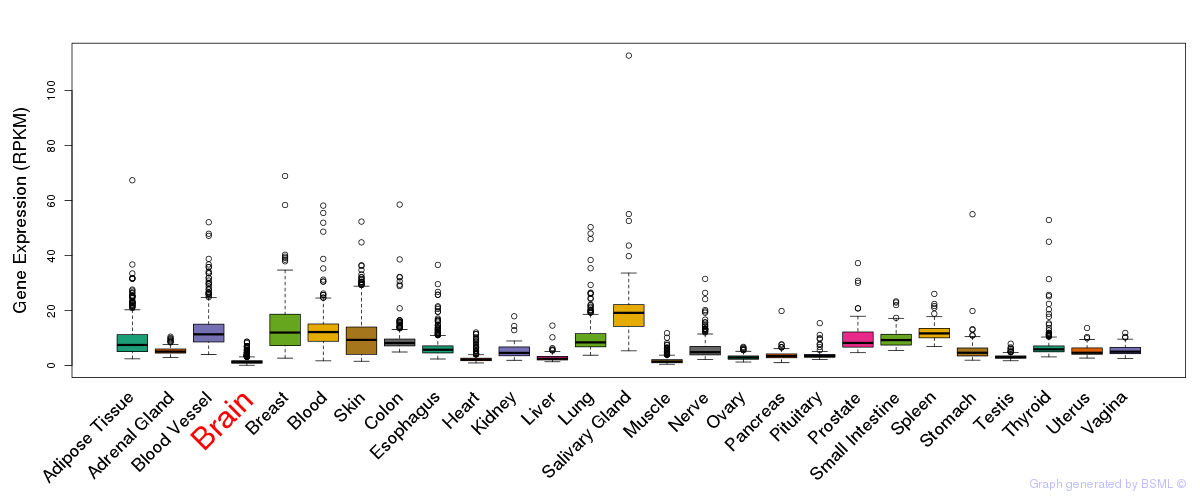
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| BBS7 | 0.84 | 0.83 |
| SACM1L | 0.84 | 0.82 |
| VPS35 | 0.84 | 0.89 |
| INSIG2 | 0.83 | 0.82 |
| GFM1 | 0.83 | 0.83 |
| NARS | 0.83 | 0.83 |
| TAX1BP1 | 0.82 | 0.85 |
| TUBGCP5 | 0.82 | 0.82 |
| RSRC1 | 0.82 | 0.79 |
| TRIM37 | 0.82 | 0.79 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.21 | -0.54 | -0.55 |
| AF347015.2 | -0.54 | -0.56 |
| AF347015.31 | -0.53 | -0.58 |
| AF347015.8 | -0.52 | -0.57 |
| MT-CO2 | -0.52 | -0.58 |
| MT-CYB | -0.51 | -0.56 |
| AF347015.18 | -0.50 | -0.57 |
| C16orf74 | -0.50 | -0.59 |
| EIF4EBP3 | -0.50 | -0.56 |
| HIGD1B | -0.49 | -0.53 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ACTA1 | ACTA | ASMA | CFTD | CFTD1 | CFTDM | MPFD | NEM1 | NEM2 | NEM3 | actin, alpha 1, skeletal muscle | Co-localization | BioGRID | 9695962 |
| CRKL | - | v-crk sarcoma virus CT10 oncogene homolog (avian)-like | Affinity Capture-Western | BioGRID | 15143164 |
| ETV6 | TEL | TEL/ABL | ets variant 6 | Affinity Capture-Western | BioGRID | 9651344 |9695962 |
| FLI1 | EWSR2 | SIC-1 | Friend leukemia virus integration 1 | - | HPRD,BioGRID | 9651344 |
| GAB2 | KIAA0571 | GRB2-associated binding protein 2 | Affinity Capture-Western | BioGRID | 15143164 |
| GRB2 | ASH | EGFRBP-GRB2 | Grb3-3 | MST084 | MSTP084 | growth factor receptor-bound protein 2 | Affinity Capture-Western Far Western | BioGRID | 15143164 |
| HDAC9 | DKFZp779K1053 | HD7 | HDAC | HDAC7 | HDAC7B | HDAC9B | HDAC9FL | HDRP | KIAA0744 | MITR | histone deacetylase 9 | Affinity Capture-Western | BioGRID | 12590135 |
| KAT5 | ESA1 | HTATIP | HTATIP1 | PLIP | TIP | TIP60 | cPLA2 | K(lysine) acetyltransferase 5 | - | HPRD,BioGRID | 12737628 |
| L3MBTL | DKFZp586P1522 | FLJ41181 | H-L(3)MBT | KIAA0681 | L3MBTL1 | dJ138B7.3 | l(3)mbt-like (Drosophila) | - | HPRD | 12588862 |
| SIN3A | DKFZp434K2235 | FLJ90319 | KIAA0700 | SIN3 homolog A, transcription regulator (yeast) | - | HPRD | 12527908 |
| UBE2I | C358B7.1 | P18 | UBC9 | ubiquitin-conjugating enzyme E2I (UBC9 homolog, yeast) | - | HPRD | 10377438 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG DORSO VENTRAL AXIS FORMATION | 25 | 21 | All SZGR 2.0 genes in this pathway |
| KORKOLA EMBRYONAL CARCINOMA UP | 41 | 27 | All SZGR 2.0 genes in this pathway |
| KORKOLA TERATOMA UP | 16 | 11 | All SZGR 2.0 genes in this pathway |
| LIU SOX4 TARGETS DN | 309 | 191 | All SZGR 2.0 genes in this pathway |
| BERTUCCI MEDULLARY VS DUCTAL BREAST CANCER UP | 206 | 111 | All SZGR 2.0 genes in this pathway |
| SENGUPTA NASOPHARYNGEAL CARCINOMA WITH LMP1 UP | 408 | 247 | All SZGR 2.0 genes in this pathway |
| WANG LMO4 TARGETS UP | 372 | 227 | All SZGR 2.0 genes in this pathway |
| JAATINEN HEMATOPOIETIC STEM CELL UP | 316 | 190 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA DN | 1375 | 806 | All SZGR 2.0 genes in this pathway |
| MARTIN INTERACT WITH HDAC | 44 | 31 | All SZGR 2.0 genes in this pathway |
| KIM MYC AMPLIFICATION TARGETS DN | 97 | 51 | All SZGR 2.0 genes in this pathway |
| PUJANA ATM PCC NETWORK | 1442 | 892 | All SZGR 2.0 genes in this pathway |
| BENPORATH ES CORE NINE CORRELATED | 100 | 68 | All SZGR 2.0 genes in this pathway |
| SMITH TERT TARGETS UP | 145 | 91 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 18HR DN | 178 | 121 | All SZGR 2.0 genes in this pathway |
| YIH RESPONSE TO ARSENITE C4 | 18 | 12 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS STEM CELL AND PROGENITOR | 681 | 420 | All SZGR 2.0 genes in this pathway |
| SAFFORD T LYMPHOCYTE ANERGY | 87 | 54 | All SZGR 2.0 genes in this pathway |
| WANG SMARCE1 TARGETS UP | 280 | 183 | All SZGR 2.0 genes in this pathway |
| MONNIER POSTRADIATION TUMOR ESCAPE UP | 393 | 244 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 3 | 720 | 440 | All SZGR 2.0 genes in this pathway |
| FOSTER TOLERANT MACROPHAGE DN | 409 | 268 | All SZGR 2.0 genes in this pathway |
| RIZKI TUMOR INVASIVENESS 3D UP | 210 | 124 | All SZGR 2.0 genes in this pathway |
| GRESHOCK CANCER COPY NUMBER UP | 323 | 240 | All SZGR 2.0 genes in this pathway |
| YAGI AML WITH T 8 21 TRANSLOCATION | 368 | 247 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS DN | 1972 | 1213 | All SZGR 2.0 genes in this pathway |
| MIYAGAWA TARGETS OF EWSR1 ETS FUSIONS UP | 259 | 159 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 1ST EGF PULSE ONLY | 1839 | 928 | All SZGR 2.0 genes in this pathway |