Gene Page: F8
Summary ?
| GeneID | 2157 |
| Symbol | F8 |
| Synonyms | AHF|DXS1253E|F8B|F8C|FVIII|HEMA |
| Description | coagulation factor VIII |
| Reference | MIM:300841|HGNC:HGNC:3546|Ensembl:ENSG00000185010|HPRD:02384|Vega:OTTHUMG00000022688 |
| Gene type | protein-coding |
| Map location | Xq28 |
| Sherlock p-value | 0.001 |
| Fetal beta | -2.052 |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenics,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
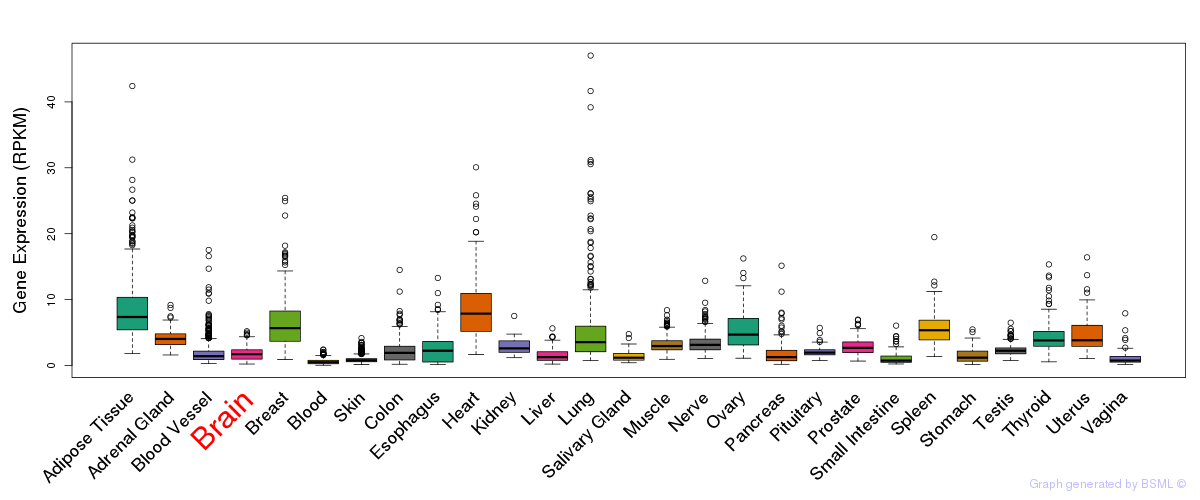
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| C11orf1 | 0.64 | 0.70 |
| GOLT1A | 0.63 | 0.48 |
| BOLA3 | 0.59 | 0.55 |
| TMPRSS6 | 0.58 | 0.36 |
| PCP4 | 0.58 | 0.54 |
| RP11-529I10.1 | 0.57 | 0.59 |
| DTNBP1 | 0.56 | 0.55 |
| C15orf24 | 0.56 | 0.60 |
| NDUFA13 | 0.56 | 0.58 |
| RND2 | 0.56 | 0.43 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| PTPRK | -0.31 | -0.32 |
| LHX2 | -0.30 | -0.39 |
| PTPN13 | -0.30 | -0.31 |
| BAT2D1 | -0.30 | -0.31 |
| HIVEP1 | -0.30 | -0.25 |
| ARHGAP21 | -0.29 | -0.25 |
| CUX2 | -0.29 | -0.25 |
| KIAA1239 | -0.29 | -0.23 |
| SRGAP3 | -0.28 | -0.24 |
| RP11-791G16.2 | -0.28 | -0.21 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| CALR | CRT | FLJ26680 | RO | SSA | cC1qR | calreticulin | - | HPRD | 9525969 |
| CANX | CNX | FLJ26570 | IP90 | P90 | calnexin | - | HPRD,BioGRID | 9525969 |
| F10 | FX | FXA | coagulation factor X | - | HPRD,BioGRID | 9759621 |10521497 |
| F9 | FIX | HEMB | MGC129641 | MGC129642 | PTC | coagulation factor IX | - | HPRD | 11278963|11830468 |
| HSPA5 | BIP | FLJ26106 | GRP78 | MIF2 | heat shock 70kDa protein 5 (glucose-regulated protein, 78kDa) | - | HPRD | 9305856 |
| LMAN1 | ERGIC-53 | ERGIC53 | F5F8D | FMFD1 | MCFD1 | MR60 | gp58 | lectin, mannose-binding, 1 | - | HPRD,BioGRID | 14629470 |
| LRP1 | A2MR | APOER | APR | CD91 | FLJ16451 | IGFBP3R | LRP | MGC88725 | TGFBR5 | low density lipoprotein-related protein 1 (alpha-2-macroglobulin receptor) | - | HPRD,BioGRID | 12522143 |
| PHYH | LN1 | LNAP1 | PAHX | PHYH1 | RD | phytanoyl-CoA 2-hydroxylase | - | HPRD,BioGRID | 11574539 |
| PROC | PC | PROC1 | protein C (inactivator of coagulation factors Va and VIIIa) | - | HPRD | 1761551 |1939075 |
| PROS1 | PROS | PS21 | PS22 | PS23 | PS24 | PS25 | PSA | protein S (alpha) | - | HPRD,BioGRID | 7620160 |
| VWF | F8VWF | VWD | von Willebrand factor | - | HPRD,BioGRID | 9218428 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG COMPLEMENT AND COAGULATION CASCADES | 69 | 49 | All SZGR 2.0 genes in this pathway |
| BIOCARTA INTRINSIC PATHWAY | 23 | 15 | All SZGR 2.0 genes in this pathway |
| REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | 89 | 56 | All SZGR 2.0 genes in this pathway |
| REACTOME FORMATION OF FIBRIN CLOT CLOTTING CASCADE | 32 | 22 | All SZGR 2.0 genes in this pathway |
| REACTOME INTRINSIC PATHWAY | 17 | 12 | All SZGR 2.0 genes in this pathway |
| REACTOME HEMOSTASIS | 466 | 331 | All SZGR 2.0 genes in this pathway |
| REACTOME PLATELET ACTIVATION SIGNALING AND AGGREGATION | 208 | 138 | All SZGR 2.0 genes in this pathway |
| GINESTIER BREAST CANCER ZNF217 AMPLIFIED UP | 78 | 48 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC1 AND HDAC2 TARGETS DN | 232 | 139 | All SZGR 2.0 genes in this pathway |
| PEREZ TP53 TARGETS | 1174 | 695 | All SZGR 2.0 genes in this pathway |
| MANALO HYPOXIA UP | 207 | 145 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE DN | 1237 | 837 | All SZGR 2.0 genes in this pathway |
| MAHAJAN RESPONSE TO IL1A DN | 76 | 57 | All SZGR 2.0 genes in this pathway |
| BONOME OVARIAN CANCER SURVIVAL SUBOPTIMAL DEBULKING | 510 | 309 | All SZGR 2.0 genes in this pathway |
| BOYAULT LIVER CANCER SUBCLASS G123 DN | 51 | 30 | All SZGR 2.0 genes in this pathway |
| CHIANG LIVER CANCER SUBCLASS PROLIFERATION DN | 179 | 97 | All SZGR 2.0 genes in this pathway |
| ZWANG DOWN BY 2ND EGF PULSE | 293 | 119 | All SZGR 2.0 genes in this pathway |