Gene Page: FANCC
Summary ?
| GeneID | 2176 |
| Symbol | FANCC |
| Synonyms | FA3|FAC|FACC |
| Description | Fanconi anemia complementation group C |
| Reference | MIM:613899|HGNC:HGNC:3584|Ensembl:ENSG00000158169|HPRD:01967|Vega:OTTHUMG00000020279 |
| Gene type | protein-coding |
| Map location | 9q22.3 |
| Pascal p-value | 0.406 |
| Sherlock p-value | 0.344 |
| Fetal beta | 1.794 |
| DMG | 2 (# studies) |
| eGene | Myers' cis & trans |
| Support | Ascano FMRP targets |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 3 |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 3 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg14115740 | 9 | 98054883 | FANCC | 4.734E-4 | 0.618 | 0.046 | DMG:Wockner_2014 |
| cg02466749 | 9 | 98054877 | FANCC | 5.575E-4 | 0.336 | 0.049 | DMG:Wockner_2014 |
| cg06202984 | 9 | 98079903 | FANCC | 1.76E-8 | -0.03 | 6.36E-6 | DMG:Jaffe_2016 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs12405921 | chr1 | 206695730 | FANCC | 2176 | 0.15 | trans | ||
| rs17572651 | chr1 | 218943612 | FANCC | 2176 | 0.17 | trans | ||
| rs16829545 | chr2 | 151977407 | FANCC | 2176 | 2.939E-10 | trans | ||
| rs3845734 | chr2 | 171125572 | FANCC | 2176 | 0.04 | trans | ||
| rs7584986 | chr2 | 184111432 | FANCC | 2176 | 7.543E-8 | trans | ||
| rs7664496 | chr4 | 56252823 | FANCC | 2176 | 0.07 | trans | ||
| rs504836 | chr4 | 56285304 | FANCC | 2176 | 0.07 | trans | ||
| rs534654 | chr4 | 56290219 | FANCC | 2176 | 0.11 | trans | ||
| snp_a-2078569 | 0 | FANCC | 2176 | 0.13 | trans | |||
| rs10491487 | chr5 | 80323367 | FANCC | 2176 | 0.19 | trans | ||
| rs16890367 | chr6 | 38078448 | FANCC | 2176 | 0.07 | trans | ||
| rs7787830 | chr7 | 98797019 | FANCC | 2176 | 0.13 | trans | ||
| rs11139334 | chr9 | 84209393 | FANCC | 2176 | 0.03 | trans | ||
| rs16955618 | chr15 | 29937543 | FANCC | 2176 | 3.081E-18 | trans | ||
| rs1041786 | chr21 | 22617710 | FANCC | 2176 | 0.09 | trans |
Section II. Transcriptome annotation
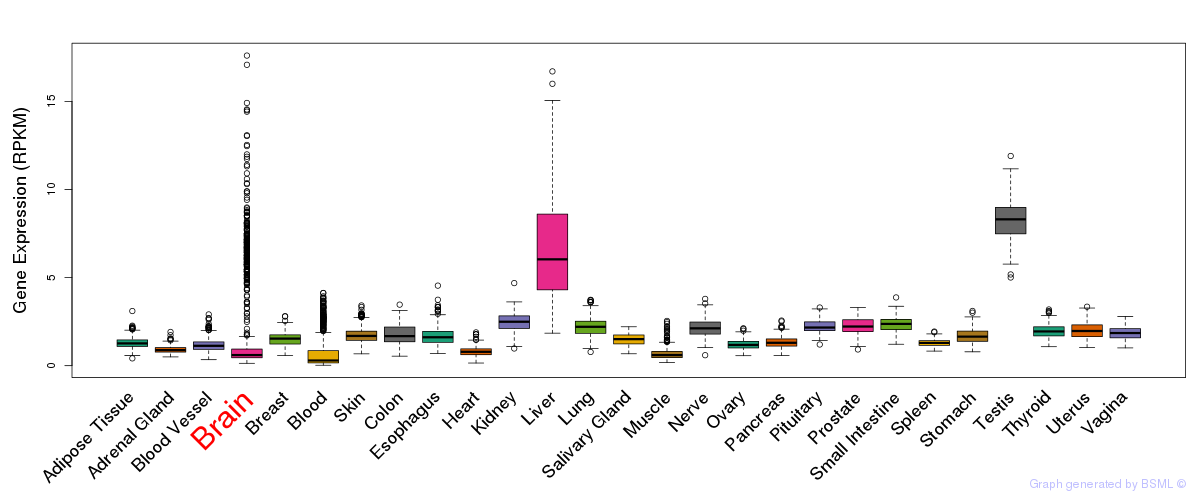
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| LUM | 0.75 | 0.54 |
| ISLR | 0.70 | 0.60 |
| ALDH1A2 | 0.70 | 0.63 |
| C7 | 0.68 | 0.54 |
| CTSK | 0.68 | 0.58 |
| COL3A1 | 0.67 | 0.20 |
| OLFML3 | 0.66 | 0.44 |
| BMP5 | 0.65 | 0.34 |
| SERPIND1 | 0.64 | 0.45 |
| OGN | 0.64 | 0.56 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.26 | -0.31 | -0.22 |
| AF347015.18 | -0.31 | -0.26 |
| MTSS1L | -0.29 | -0.34 |
| SMOX | -0.29 | -0.33 |
| AF347015.2 | -0.28 | -0.18 |
| AF347015.15 | -0.27 | -0.18 |
| AF347015.8 | -0.27 | -0.18 |
| AC010300.1 | -0.27 | -0.29 |
| MMD2 | -0.26 | -0.23 |
| AF347015.33 | -0.26 | -0.17 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ATP6 | MTATP6 | ATP synthase 6; ATPase subunit 6 | Two-hybrid | BioGRID | 14499622 |
| AZIN1 | MGC3832 | MGC691 | OAZI | OAZIN | ODC1L | antizyme inhibitor 1 | Two-hybrid | BioGRID | 14499622 |
| CDC2 | CDC28A | CDK1 | DKFZp686L20222 | MGC111195 | cell division cycle 2, G1 to S and G2 to M | - | HPRD,BioGRID | 9242535 |
| EIF2AK2 | EIF2AK1 | MGC126524 | PKR | PRKR | eukaryotic translation initiation factor 2-alpha kinase 2 | Affinity Capture-Western | BioGRID | 12397061 |
| FANCA | FA | FA-H | FA1 | FAA | FACA | FAH | FANCH | MGC75158 | Fanconi anemia, complementation group A | Affinity Capture-MS Affinity Capture-Western Reconstituted Complex Two-hybrid | BioGRID | 10373536 |10551855 |10627486 |10961856 |11063725 |12210728 |12239156 |12973351 |15082718 |
| FANCA | FA | FA-H | FA1 | FAA | FACA | FAH | FANCH | MGC75158 | Fanconi anemia, complementation group A | - | HPRD | 9742112 |
| FANCD2 | DKFZp762A223 | FA-D2 | FA4 | FACD | FAD | FAD2 | FANCD | FLJ23826 | Fanconi anemia, complementation group D2 | - | HPRD | 11751423 |
| FANCE | FACE | FAE | Fanconi anemia, complementation group E | - | HPRD,BioGRID | 11157805 |
| FANCF | FAF | MGC126856 | Fanconi anemia, complementation group F | - | HPRD,BioGRID | 11063725 |
| FANCG | FAG | XRCC9 | Fanconi anemia, complementation group G | - | HPRD,BioGRID | 10373536 |
| FLJ10213 | - | hypothetical protein FLJ10213 | Affinity Capture-MS | BioGRID | 17353931 |
| GSTP1 | DFN7 | FAEES3 | GST3 | PI | glutathione S-transferase pi 1 | Affinity Capture-Western Two-hybrid | BioGRID | 11433346 |14499622 |
| HSP90AA1 | FLJ31884 | HSP86 | HSP89A | HSP90A | HSP90N | HSPC1 | HSPCA | HSPCAL1 | HSPCAL4 | HSPN | Hsp89 | Hsp90 | LAP2 | heat shock protein 90kDa alpha (cytosolic), class A member 1 | Two-hybrid | BioGRID | 14499622 |
| HSP90B1 | ECGP | GP96 | GRP94 | TRA1 | heat shock protein 90kDa beta (Grp94), member 1 | Two-hybrid | BioGRID | 14499622 |
| HSPA1A | FLJ54303 | FLJ54370 | FLJ54392 | FLJ54408 | FLJ75127 | HSP70-1 | HSP70-1A | HSP70I | HSP72 | HSPA1 | HSPA1B | heat shock 70kDa protein 1A | Affinity Capture-Western Two-hybrid | BioGRID | 12397061 |14499622 |
| HSPA1A | FLJ54303 | FLJ54370 | FLJ54392 | FLJ54408 | FLJ75127 | HSP70-1 | HSP70-1A | HSP70I | HSP72 | HSPA1 | HSPA1B | heat shock 70kDa protein 1A | - | HPRD | 11500375 |12397061 |
| HSPA8 | HSC54 | HSC70 | HSC71 | HSP71 | HSP73 | HSPA10 | LAP1 | MGC131511 | MGC29929 | NIP71 | heat shock 70kDa protein 8 | Two-hybrid | BioGRID | 14499622 |
| IK | CSA2 | MGC59741 | RED | IK cytokine, down-regulator of HLA II | Two-hybrid | BioGRID | 14499622 |
| KRT1 | CK1 | EHK1 | K1 | KRT1A | keratin 1 | Two-hybrid | BioGRID | 14499622 |
| RPL18 | - | ribosomal protein L18 | Two-hybrid | BioGRID | 14499622 |
| RPS3A | FTE1 | MFTL | MGC23240 | ribosomal protein S3A | Two-hybrid | BioGRID | 14499622 |
| SPTA1 | EL2 | SPTA | spectrin, alpha, erythrocytic 1 (elliptocytosis 2) | - | HPRD | 10551855 |
| SPTAN1 | (ALPHA)II-SPECTRIN | FLJ44613 | NEAS | spectrin, alpha, non-erythrocytic 1 (alpha-fodrin) | Affinity Capture-Western Co-purification | BioGRID | 10551855 |11401546 |
| STAT1 | DKFZp686B04100 | ISGF-3 | STAT91 | signal transducer and activator of transcription 1, 91kDa | - | HPRD,BioGRID | 10848598 |
| TCERG1 | CA150 | MGC133200 | TAF2S | transcription elongation regulator 1 | Two-hybrid | BioGRID | 14499622 |
| USP14 | TGT | ubiquitin specific peptidase 14 (tRNA-guanine transglycosylase) | Two-hybrid | BioGRID | 14499622 |
| ZBTB32 | FAXF | FAZF | Rog | TZFP | ZNF538 | zinc finger and BTB domain containing 32 | Affinity Capture-Western Two-hybrid | BioGRID | 10572087 |14499622 |
| ZBTB32 | FAXF | FAZF | Rog | TZFP | ZNF538 | zinc finger and BTB domain containing 32 | - | HPRD | 10572087 |11986317 |
| ZBTB32 | FAXF | FAZF | Rog | TZFP | ZNF538 | zinc finger and BTB domain containing 32 | - | HPRD | 11986317 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| BIOCARTA ATRBRCA PATHWAY | 21 | 12 | All SZGR 2.0 genes in this pathway |
| PID FANCONI PATHWAY | 47 | 28 | All SZGR 2.0 genes in this pathway |
| PID BARD1 PATHWAY | 29 | 19 | All SZGR 2.0 genes in this pathway |
| REACTOME FANCONI ANEMIA PATHWAY | 25 | 11 | All SZGR 2.0 genes in this pathway |
| REACTOME DNA REPAIR | 112 | 59 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN DN | 1781 | 1082 | All SZGR 2.0 genes in this pathway |
| MATHEW FANCONI ANEMIA GENES | 11 | 7 | All SZGR 2.0 genes in this pathway |
| MYLLYKANGAS AMPLIFICATION HOT SPOT 8 | 18 | 7 | All SZGR 2.0 genes in this pathway |
| SCHLOSSER SERUM RESPONSE DN | 712 | 443 | All SZGR 2.0 genes in this pathway |
| MOHANKUMAR TLX1 TARGETS UP | 414 | 287 | All SZGR 2.0 genes in this pathway |
| KIM MYCN AMPLIFICATION TARGETS UP | 92 | 64 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA1 PCC NETWORK | 1652 | 1023 | All SZGR 2.0 genes in this pathway |
| LOPEZ MBD TARGETS | 957 | 597 | All SZGR 2.0 genes in this pathway |
| WEI MYCN TARGETS WITH E BOX | 795 | 478 | All SZGR 2.0 genes in this pathway |
| BENPORATH SOX2 TARGETS | 734 | 436 | All SZGR 2.0 genes in this pathway |
| MORI LARGE PRE BII LYMPHOCYTE UP | 86 | 49 | All SZGR 2.0 genes in this pathway |
| KAUFFMANN DNA REPAIR GENES | 230 | 137 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS STEM CELL AND PROGENITOR | 681 | 420 | All SZGR 2.0 genes in this pathway |
| FOSTER TOLERANT MACROPHAGE DN | 409 | 268 | All SZGR 2.0 genes in this pathway |
| MATZUK MEIOTIC AND DNA REPAIR | 39 | 26 | All SZGR 2.0 genes in this pathway |
| GRESHOCK CANCER COPY NUMBER UP | 323 | 240 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS UP | 1305 | 895 | All SZGR 2.0 genes in this pathway |
| SWEET LUNG CANCER KRAS DN | 435 | 289 | All SZGR 2.0 genes in this pathway |
| DUTERTRE ESTRADIOL RESPONSE 24HR UP | 324 | 193 | All SZGR 2.0 genes in this pathway |
| WAKABAYASHI ADIPOGENESIS PPARG BOUND 8D | 658 | 397 | All SZGR 2.0 genes in this pathway |
| ROESSLER LIVER CANCER METASTASIS UP | 107 | 72 | All SZGR 2.0 genes in this pathway |