Gene Page: ALDH1B1
Summary ?
| GeneID | 219 |
| Symbol | ALDH1B1 |
| Synonyms | ALDH5|ALDHX |
| Description | aldehyde dehydrogenase 1 family member B1 |
| Reference | MIM:100670|HGNC:HGNC:407|Ensembl:ENSG00000137124|HPRD:00005|Vega:OTTHUMG00000019938 |
| Gene type | protein-coding |
| Map location | 9p11.1 |
| Pascal p-value | 0.539 |
| Fetal beta | -0.156 |
| DMG | 1 (# studies) |
| eGene | Caudate basal ganglia Nucleus accumbens basal ganglia |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg22397447 | 9 | 38392975 | ALDH1B1 | 6.46E-9 | -0.01 | 3.36E-6 | DMG:Jaffe_2016 |
Section II. Transcriptome annotation
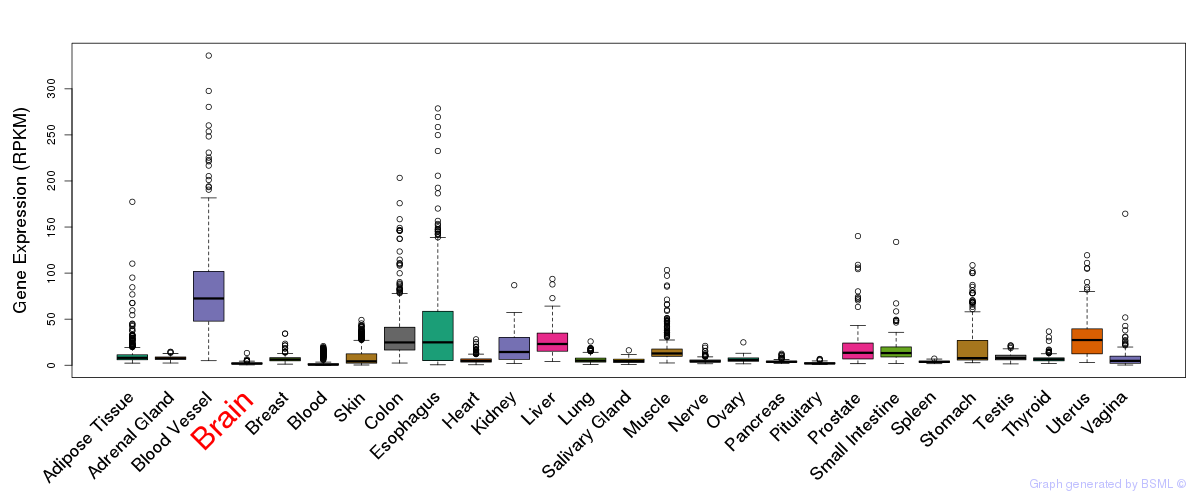
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG GLYCOLYSIS GLUCONEOGENESIS | 62 | 41 | All SZGR 2.0 genes in this pathway |
| KEGG ASCORBATE AND ALDARATE METABOLISM | 25 | 17 | All SZGR 2.0 genes in this pathway |
| KEGG FATTY ACID METABOLISM | 42 | 29 | All SZGR 2.0 genes in this pathway |
| KEGG VALINE LEUCINE AND ISOLEUCINE DEGRADATION | 44 | 26 | All SZGR 2.0 genes in this pathway |
| KEGG LYSINE DEGRADATION | 44 | 29 | All SZGR 2.0 genes in this pathway |
| KEGG ARGININE AND PROLINE METABOLISM | 54 | 39 | All SZGR 2.0 genes in this pathway |
| KEGG HISTIDINE METABOLISM | 29 | 19 | All SZGR 2.0 genes in this pathway |
| KEGG TRYPTOPHAN METABOLISM | 40 | 33 | All SZGR 2.0 genes in this pathway |
| KEGG BETA ALANINE METABOLISM | 22 | 16 | All SZGR 2.0 genes in this pathway |
| KEGG GLYCEROLIPID METABOLISM | 49 | 26 | All SZGR 2.0 genes in this pathway |
| KEGG PYRUVATE METABOLISM | 40 | 26 | All SZGR 2.0 genes in this pathway |
| KEGG PROPANOATE METABOLISM | 33 | 22 | All SZGR 2.0 genes in this pathway |
| KEGG BUTANOATE METABOLISM | 34 | 20 | All SZGR 2.0 genes in this pathway |
| KEGG LIMONENE AND PINENE DEGRADATION | 10 | 8 | All SZGR 2.0 genes in this pathway |
| COLDREN GEFITINIB RESISTANCE UP | 85 | 57 | All SZGR 2.0 genes in this pathway |
| DELYS THYROID CANCER DN | 232 | 154 | All SZGR 2.0 genes in this pathway |
| VANHARANTA UTERINE FIBROID WITH 7Q DELETION DN | 36 | 23 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN DN | 1781 | 1082 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 XPCS UP | 28 | 16 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 UP | 309 | 199 | All SZGR 2.0 genes in this pathway |
| FURUKAWA DUSP6 TARGETS PCI35 UP | 74 | 32 | All SZGR 2.0 genes in this pathway |
| BUYTAERT PHOTODYNAMIC THERAPY STRESS DN | 637 | 377 | All SZGR 2.0 genes in this pathway |
| DEBIASI APOPTOSIS BY REOVIRUS INFECTION DN | 287 | 208 | All SZGR 2.0 genes in this pathway |
| NIELSEN LEIOMYOSARCOMA CNN1 UP | 19 | 13 | All SZGR 2.0 genes in this pathway |
| BILD MYC ONCOGENIC SIGNATURE | 206 | 117 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 6HR DN | 911 | 527 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 24HR DN | 1011 | 592 | All SZGR 2.0 genes in this pathway |
| OUILLETTE CLL 13Q14 DELETION DN | 60 | 38 | All SZGR 2.0 genes in this pathway |
| CHEN METABOLIC SYNDROM NETWORK | 1210 | 725 | All SZGR 2.0 genes in this pathway |
| MEISSNER NPC HCP WITH H3 UNMETHYLATED | 536 | 296 | All SZGR 2.0 genes in this pathway |
| HOSHIDA LIVER CANCER SUBCLASS S3 | 266 | 180 | All SZGR 2.0 genes in this pathway |
| CAIRO LIVER DEVELOPMENT DN | 222 | 141 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 4HR DN | 254 | 158 | All SZGR 2.0 genes in this pathway |
| MARTENS TRETINOIN RESPONSE UP | 857 | 456 | All SZGR 2.0 genes in this pathway |
| WANG RESPONSE TO GSK3 INHIBITOR SB216763 DN | 374 | 217 | All SZGR 2.0 genes in this pathway |
| PANGAS TUMOR SUPPRESSION BY SMAD1 AND SMAD5 UP | 134 | 85 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE VIA TP53 GROUP D | 280 | 158 | All SZGR 2.0 genes in this pathway |
| PURBEY TARGETS OF CTBP1 AND SATB1 DN | 180 | 116 | All SZGR 2.0 genes in this pathway |
| PURBEY TARGETS OF CTBP1 NOT SATB1 UP | 344 | 215 | All SZGR 2.0 genes in this pathway |