Gene Page: MED19
Summary ?
| GeneID | 219541 |
| Symbol | MED19 |
| Synonyms | DT2P1G7|LCMR1|MED19AS |
| Description | mediator complex subunit 19 |
| Reference | MIM:612385|HGNC:HGNC:29600|Ensembl:ENSG00000156603|HPRD:17477|Vega:OTTHUMG00000167199 |
| Gene type | protein-coding |
| Map location | 11q12.1 |
| Pascal p-value | 3.987E-7 |
| Sherlock p-value | 0.196 |
| Fetal beta | 0.499 |
| DMG | 1 (# studies) |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Nishioka_2013 | Genome-wide DNA methylation analysis | The authors investigated the methylation profiles of DNA in peripheral blood cells from 18 patients with first-episode schizophrenia (FESZ) and from 15 normal controls. | 1 |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.034 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg27497687 | 11 | 57479637 | MED19 | -0.026 | 0.25 | DMG:Nishioka_2013 |
Section II. Transcriptome annotation
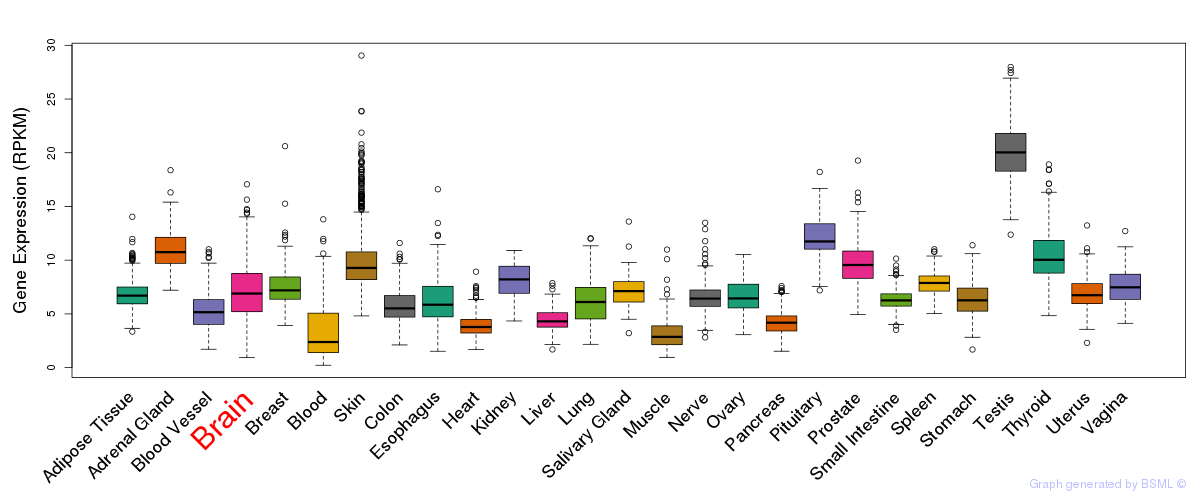
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005515 | protein binding | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006355 | regulation of transcription, DNA-dependent | IEA | - | |
| GO:0006350 | transcription | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0000119 | mediator complex | IEA | - | |
| GO:0005634 | nucleus | IEA | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| CCNC | CycC | cyclin C | Affinity Capture-MS | BioGRID | 15175163 |
| CDC2L6 | CDK11 | KIAA1028 | bA346C16.3 | cell division cycle 2-like 6 (CDK8-like) | Affinity Capture-MS | BioGRID | 15175163 |
| CDK8 | K35 | MGC126074 | MGC126075 | cyclin-dependent kinase 8 | Affinity Capture-MS | BioGRID | 15175163 |
| DCK | MGC117410 | MGC138632 | deoxycytidine kinase | Two-hybrid | BioGRID | 16169070 |
| MED1 | CRSP1 | CRSP200 | DRIP205 | DRIP230 | MGC71488 | PBP | PPARBP | PPARGBP | RB18A | TRAP220 | TRIP2 | mediator complex subunit 1 | Affinity Capture-MS Affinity Capture-Western | BioGRID | 14638676 |15175163 |
| MED10 | L6 | MGC5309 | NUT2 | TRG20 | mediator complex subunit 10 | Affinity Capture-MS Affinity Capture-Western | BioGRID | 12584197 |14638676 |15175163 |
| MED11 | HSPC296 | MGC88387 | mediator complex subunit 11 | Affinity Capture-MS Affinity Capture-Western | BioGRID | 14576168 |14638676 |15175163 |
| MED12 | CAGH45 | HOPA | KIAA0192 | OPA1 | TNRC11 | TRAP230 | mediator complex subunit 12 | Affinity Capture-MS | BioGRID | 15175163 |
| MED13 | ARC250 | DRIP250 | HSPC221 | KIAA0593 | THRAP1 | TRAP240 | mediator complex subunit 13 | Affinity Capture-MS | BioGRID | 15175163 |
| MED13L | DKFZp781D0112 | FLJ21627 | KIAA1025 | PROSIT240 | THRAP2 | TRAP240L | mediator complex subunit 13-like | Affinity Capture-MS | BioGRID | 15175163 |
| MED14 | CRSP150 | CRSP2 | CSRP | CXorf4 | DRIP150 | EXLM1 | MGC104513 | RGR1 | TRAP170 | mediator complex subunit 14 | Affinity Capture-MS | BioGRID | 15175163 |
| MED15 | ARC105 | CAG7A | CTG7A | DKFZp686A2214 | DKFZp762B1216 | FLJ42282 | FLJ42935 | PCQAP | TIG-1 | TIG1 | TNRC7 | mediator complex subunit 15 | Affinity Capture-MS | BioGRID | 15175163 |
| MED16 | DRIP92 | THRAP5 | TRAP95 | mediator complex subunit 16 | Affinity Capture-MS | BioGRID | 15175163 |
| MED17 | CRSP6 | CRSP77 | DRIP80 | FLJ10812 | TRAP80 | mediator complex subunit 17 | Affinity Capture-MS Affinity Capture-Western Reconstituted Complex | BioGRID | 12584197 |14576168 |14638676 |15175163 |
| MED17 | CRSP6 | CRSP77 | DRIP80 | FLJ10812 | TRAP80 | mediator complex subunit 17 | TRAP80 interacts with LCMR1. This interaction was modelled on a demonstrated interaction between human TRAP80 and mouse LCMR1. | BIND | 12584197 |
| MED18 | FLJ20045 | p28b | mediator complex subunit 18 | Affinity Capture-MS Affinity Capture-Western | BioGRID | 14576168 |14638676 |15175163 |
| MED19 | DT2P1G7 | LCMR1 | mediator complex subunit 19 | Affinity Capture-MS Affinity Capture-Western | BioGRID | 14638676 |15175163 |
| MED20 | DKFZp586D2223 | MGC29869 | PRO0213 | TRFP | mediator complex subunit 20 | Affinity Capture-MS Affinity Capture-Western | BioGRID | 14576168 |15175163 |
| MED21 | SRB7 | SURB7 | mediator complex subunit 21 | Affinity Capture-MS | BioGRID | 15175163 |
| MED22 | MED24 | MGC48682 | SURF5 | mediator complex subunit 22 | Affinity Capture-MS | BioGRID | 15175163 |
| MED23 | CRSP130 | CRSP133 | CRSP3 | DKFZp434H0117 | DRIP130 | SUR2 | mediator complex subunit 23 | Affinity Capture-MS | BioGRID | 15175163 |
| MED24 | ARC100 | CRSP100 | CRSP4 | DRIP100 | KIAA0130 | MGC8748 | THRAP4 | TRAP100 | mediator complex subunit 24 | Affinity Capture-MS | BioGRID | 15175163 |
| MED25 | ACID1 | ARC92 | DKFZp434K0512 | MGC70671 | P78 | PTOV2 | TCBAP0758 | mediator complex subunit 25 | Affinity Capture-MS Affinity Capture-Western | BioGRID | 14638676 |15175163 |
| MED26 | CRSP7 | CRSP70 | mediator complex subunit 26 | Affinity Capture-MS | BioGRID | 15175163 |
| MED27 | CRAP34 | CRSP34 | CRSP8 | MGC11274 | TRAP37 | mediator complex subunit 27 | TRAP37 interacts with LCMR1. This interaction was modelled on a demonstrated interaction between human TRAP37 and mouse LCMR1. | BIND | 12584197 |
| MED27 | CRAP34 | CRSP34 | CRSP8 | MGC11274 | TRAP37 | mediator complex subunit 27 | Affinity Capture-MS Reconstituted Complex | BioGRID | 12584197 |15175163 |
| MED28 | 1500003D12Rik | DKFZp434N185 | EG1 | magicin | mediator complex subunit 28 | - | HPRD,BioGRID | 15175163 |
| MED29 | DKFZp434H247 | IXL | mediator complex subunit 29 | - | HPRD,BioGRID | 14576168 |
| MED30 | MGC9890 | THRAP6 | TRAP25 | mediator complex subunit 30 | Affinity Capture-MS | BioGRID | 15175163 |
| MED31 | 3110004H13Rik | CGI-125 | FLJ27436 | FLJ36714 | Soh1 | mediator complex subunit 31 | - | HPRD,BioGRID | 12584197 |
| MED4 | DRIP36 | FLJ10956 | HSPC126 | RP11-90M2.2 | TRAP36 | VDRIP | mediator complex subunit 4 | Affinity Capture-MS Affinity Capture-Western | BioGRID | 14638676 |15175163 |
| MED6 | NY-REN-28 | mediator complex subunit 6 | Affinity Capture-MS Affinity Capture-Western | BioGRID | 14576168 |15175163 |
| MED7 | CRSP33 | CRSP9 | MGC12284 | mediator complex subunit 7 | Affinity Capture-MS | BioGRID | 15175163 |
| MED8 | ARC32 | MGC17544 | MGC19641 | mediator complex subunit 8 | Affinity Capture-MS Affinity Capture-Western | BioGRID | 14576168 |14638676 |15175163 |
| MED9 | FLJ10193 | MED25 | MGC138234 | mediator complex subunit 9 | Affinity Capture-MS | BioGRID | 15175163 |
| MED9 | FLJ10193 | MED25 | MGC138234 | mediator complex subunit 9 | - | HPRD | 14638676 |
| NFKBIA | IKBA | MAD-3 | NFKBI | nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha | Two-hybrid | BioGRID | 16169070 |
| POLR2A | MGC75453 | POLR2 | POLRA | RPB1 | RPBh1 | RPO2 | RPOL2 | RpIILS | hRPB220 | hsRPB1 | polymerase (RNA) II (DNA directed) polypeptide A, 220kDa | Affinity Capture-MS | BioGRID | 15175163 |
| POLR2B | POL2RB | RPB2 | hRPB140 | hsRPB2 | polymerase (RNA) II (DNA directed) polypeptide B, 140kDa | Affinity Capture-MS | BioGRID | 15175163 |
| POLR2C | RPB3 | RPB31 | hRPB33 | hsRPB3 | polymerase (RNA) II (DNA directed) polypeptide C, 33kDa | Affinity Capture-MS | BioGRID | 15175163 |
| POLR2D | HSRBP4 | HSRPB4 | RBP4 | polymerase (RNA) II (DNA directed) polypeptide D | Affinity Capture-MS | BioGRID | 15175163 |
| POLR2E | RPABC1 | RPB5 | XAP4 | hRPB25 | hsRPB5 | polymerase (RNA) II (DNA directed) polypeptide E, 25kDa | Affinity Capture-MS | BioGRID | 15175163 |
| POLR2F | HRBP14.4 | POLRF | RPABC2 | RPB14.4 | RPB6 | polymerase (RNA) II (DNA directed) polypeptide F | Affinity Capture-MS | BioGRID | 15175163 |
| POLR2G | MGC138367 | MGC138369 | RPB7 | hRPB19 | hsRPB7 | polymerase (RNA) II (DNA directed) polypeptide G | Affinity Capture-MS | BioGRID | 15175163 |
| POLR2H | RPABC3 | RPB17 | RPB8 | hsRPB8 | polymerase (RNA) II (DNA directed) polypeptide H | Affinity Capture-MS | BioGRID | 15175163 |
| POLR2I | RPB9 | hRPB14.5 | polymerase (RNA) II (DNA directed) polypeptide I, 14.5kDa | Affinity Capture-MS | BioGRID | 15175163 |
| POLR2J | MGC71910 | POLR2J1 | RPB11 | RPB11A | RPB11m | hRPB14 | polymerase (RNA) II (DNA directed) polypeptide J, 13.3kDa | Affinity Capture-MS | BioGRID | 15175163 |
| POLR2L | RBP10 | RPABC5 | RPB10 | RPB10beta | RPB7.6 | hRPB7.6 | hsRPB10b | polymerase (RNA) II (DNA directed) polypeptide L, 7.6kDa | Affinity Capture-MS | BioGRID | 15175163 |
| RADIL | FLJ10324 | KIAA1849 | MGC161589 | Rap GTPase interactor | Two-hybrid | BioGRID | 16169070 |
| TFF1 | BCEI | D21S21 | HP1.A | HPS2 | pNR-2 | pS2 | trefoil factor 1 | MED19 interacts with pS2 promoter. | BIND | 15989967 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| REACTOME DEVELOPMENTAL BIOLOGY | 396 | 292 | All SZGR 2.0 genes in this pathway |
| REACTOME PPARA ACTIVATES GENE EXPRESSION | 104 | 72 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF LIPIDS AND LIPOPROTEINS | 478 | 302 | All SZGR 2.0 genes in this pathway |
| REACTOME FATTY ACID TRIACYLGLYCEROL AND KETONE BODY METABOLISM | 168 | 115 | All SZGR 2.0 genes in this pathway |
| REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | 72 | 53 | All SZGR 2.0 genes in this pathway |
| OSMAN BLADDER CANCER UP | 404 | 246 | All SZGR 2.0 genes in this pathway |
| SHETH LIVER CANCER VS TXNIP LOSS PAM6 | 46 | 32 | All SZGR 2.0 genes in this pathway |
| JOHNSTONE PARVB TARGETS 3 DN | 918 | 550 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-214 | 633 | 640 | 1A,m8 | hsa-miR-214brain | ACAGCAGGCACAGACAGGCAG |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.