Gene Page: ARMC3
Summary ?
| GeneID | 219681 |
| Symbol | ARMC3 |
| Synonyms | CT81|KU-CT-1 |
| Description | armadillo repeat containing 3 |
| Reference | MIM:611226|HGNC:HGNC:30964|Ensembl:ENSG00000165309|HPRD:12487|Vega:OTTHUMG00000017811 |
| Gene type | protein-coding |
| Map location | 10p12.31 |
| Pascal p-value | 0.205 |
| Fetal beta | -0.497 |
| DMG | 1 (# studies) |
| eGene | Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg16819392 | 10 | 23326418 | ARMC3 | 4.347E-4 | -0.674 | 0.045 | DMG:Wockner_2014 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs4294953 | chr2 | 154365614 | ARMC3 | 219681 | 0.16 | trans | ||
| rs9681394 | chr3 | 43256230 | ARMC3 | 219681 | 0.03 | trans | ||
| rs9882567 | chr3 | 188988929 | ARMC3 | 219681 | 0.11 | trans | ||
| rs1981925 | chr4 | 34348578 | ARMC3 | 219681 | 1.832E-5 | trans | ||
| rs2291715 | chr4 | 75485871 | ARMC3 | 219681 | 0.12 | trans | ||
| rs6836195 | chr4 | 120756689 | ARMC3 | 219681 | 0.04 | trans | ||
| rs10075513 | chr5 | 117091552 | ARMC3 | 219681 | 0.11 | trans | ||
| rs2268637 | chr6 | 39050917 | ARMC3 | 219681 | 0 | trans | ||
| rs10953482 | chr7 | 105548000 | ARMC3 | 219681 | 0.17 | trans | ||
| rs2524878 | chr7 | 125222418 | ARMC3 | 219681 | 0.09 | trans | ||
| rs2524880 | chr7 | 125222519 | ARMC3 | 219681 | 0.14 | trans | ||
| rs10958899 | chr9 | 10128700 | ARMC3 | 219681 | 0.03 | trans | ||
| rs10958907 | chr9 | 10131580 | ARMC3 | 219681 | 0.03 | trans | ||
| rs1322159 | chr9 | 10132695 | ARMC3 | 219681 | 0.02 | trans | ||
| rs10958910 | chr9 | 10134759 | ARMC3 | 219681 | 0.04 | trans | ||
| rs2140736 | chr15 | 31159211 | ARMC3 | 219681 | 0.04 | trans | ||
| rs11656351 | chr17 | 9489574 | ARMC3 | 219681 | 0.02 | trans | ||
| rs11080561 | chr18 | 12257229 | ARMC3 | 219681 | 0.1 | trans | ||
| rs16951170 | chr18 | 47402487 | ARMC3 | 219681 | 0.02 | trans | ||
| rs7268640 | chr20 | 47150267 | ARMC3 | 219681 | 1.158E-5 | trans |
Section II. Transcriptome annotation
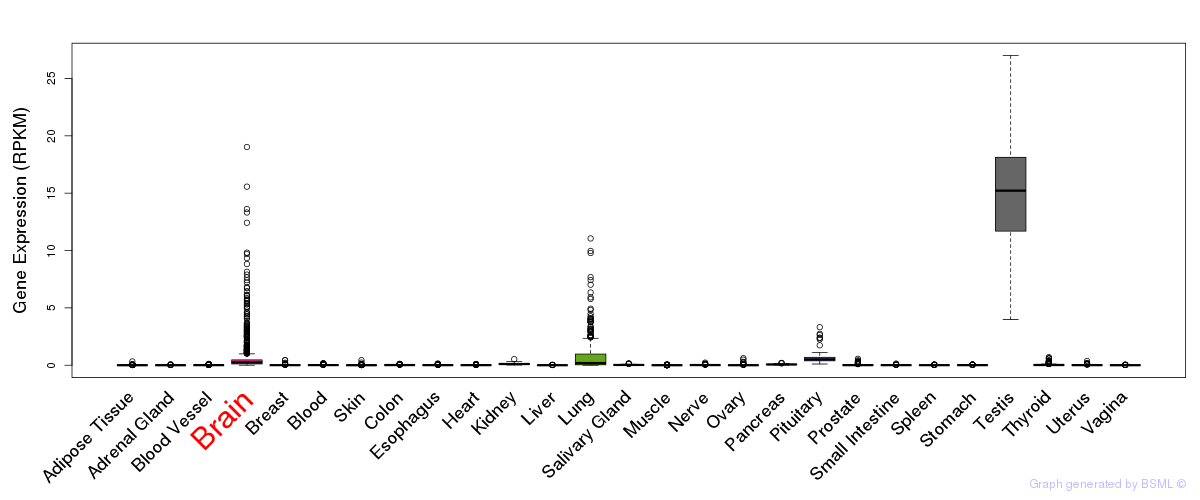
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| SENGUPTA NASOPHARYNGEAL CARCINOMA DN | 349 | 157 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA UP | 1821 | 933 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 2 | 473 | 224 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER CANCER WITH H3K9ME3 UP | 141 | 75 | All SZGR 2.0 genes in this pathway |
| ACEVEDO METHYLATED IN LIVER CANCER DN | 940 | 425 | All SZGR 2.0 genes in this pathway |
| CHEN METABOLIC SYNDROM NETWORK | 1210 | 725 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN ES ICP WITH H3K4ME3 | 718 | 401 | All SZGR 2.0 genes in this pathway |
| NOUSHMEHR GBM SILENCED BY METHYLATION | 50 | 32 | All SZGR 2.0 genes in this pathway |
| RAO BOUND BY SALL4 ISOFORM A | 182 | 108 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 1ST EGF PULSE ONLY | 1839 | 928 | All SZGR 2.0 genes in this pathway |