Gene Page: KIF6
Summary ?
| GeneID | 221458 |
| Symbol | KIF6 |
| Synonyms | C6orf102|dJ1043E3.1|dJ137F1.4|dJ188D3.1 |
| Description | kinesin family member 6 |
| Reference | MIM:613919|HGNC:HGNC:21202|Ensembl:ENSG00000164627|HPRD:10786|Vega:OTTHUMG00000014648 |
| Gene type | protein-coding |
| Map location | 6p21.2 |
| Pascal p-value | 0.603 |
| eGene | Caudate basal ganglia Cerebellum Cortex Frontal Cortex BA9 Myers' cis & trans |
| Support | CompositeSet |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DNM:Fromer_2014 | Whole Exome Sequencing analysis | This study reported a WES study of 623 schizophrenia trios, reporting DNMs using genomic DNA. | |
| GSMA_I | Genome scan meta-analysis | Psr: 0.033 | |
| GSMA_IIE | Genome scan meta-analysis (European-ancestry samples) | Psr: 0.04433 |
Section I. Genetics and epigenetics annotation
 DNM table
DNM table
| Gene | Chromosome | Position | Ref | Alt | Transcript | AA change | Mutation type | Sift | CG46 | Trait | Study |
|---|---|---|---|---|---|---|---|---|---|---|---|
| KIF6 | chr6 | 39513377 | G | C | NM_145027 | p.423H>Q | missense | Schizophrenia | DNM:Fromer_2014 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs17027208 | chr3 | 31219519 | KIF6 | 221458 | 0.15 | trans | ||
| rs7023147 | chr9 | 10077179 | KIF6 | 221458 | 0.14 | trans | ||
| rs10958896 | chr9 | 10126894 | KIF6 | 221458 | 0.15 | trans | ||
| rs10958899 | chr9 | 10128700 | KIF6 | 221458 | 0.05 | trans | ||
| rs10958907 | chr9 | 10131580 | KIF6 | 221458 | 0.08 | trans | ||
| rs1322159 | chr9 | 10132695 | KIF6 | 221458 | 0.12 | trans | ||
| rs10958910 | chr9 | 10134759 | KIF6 | 221458 | 0.12 | trans | ||
| rs10994209 | chr10 | 61877705 | KIF6 | 221458 | 0.01 | trans |
Section II. Transcriptome annotation
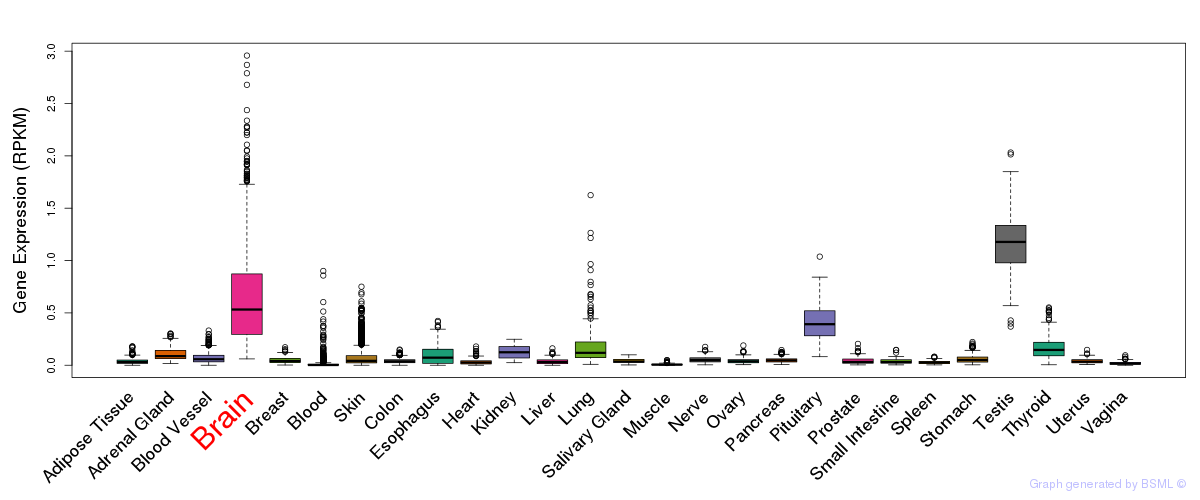
General gene expression (GTEx)
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0000166 | nucleotide binding | IEA | - | |
| GO:0003777 | microtubule motor activity | IEA | - | |
| GO:0005524 | ATP binding | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007018 | microtubule-based movement | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005874 | microtubule | IEA | - | |
| GO:0005875 | microtubule associated complex | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| DODD NASOPHARYNGEAL CARCINOMA UP | 1821 | 933 | All SZGR 2.0 genes in this pathway |
| DARWICHE PAPILLOMA PROGRESSION RISK | 74 | 44 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS STEM CELL LONG TERM | 302 | 191 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 2 | 473 | 224 | All SZGR 2.0 genes in this pathway |
| KUMAR PATHOGEN LOAD BY MACROPHAGES | 275 | 155 | All SZGR 2.0 genes in this pathway |