Gene Page: PHACTR1
Summary ?
| GeneID | 221692 |
| Symbol | PHACTR1 |
| Synonyms | RPEL|RPEL1|dJ257A7.2 |
| Description | phosphatase and actin regulator 1 |
| Reference | MIM:608723|HGNC:HGNC:20990|Ensembl:ENSG00000112137|HPRD:12283|Vega:OTTHUMG00000014270 |
| Gene type | protein-coding |
| Map location | 6p24.1 |
| Pascal p-value | 0.339 |
| Sherlock p-value | 0.718 |
| Fetal beta | -1.314 |
| DMG | 1 (# studies) |
| eGene | Myers' cis & trans |
| Support | G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-FULL G2Cdb.human_BAYES-COLLINS-MOUSE-PSD-CONSENSUS CompositeSet Darnell FMRP targets |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 1 |
| GSMA_I | Genome scan meta-analysis | Psr: 0.0159 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg21538684 | 6 | 13274180 | PHACTR1 | 2.225E-4 | 0.295 | 0.036 | DMG:Wockner_2014 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs4715861 | chr6 | 14009618 | PHACTR1 | 221692 | 0.08 | cis | ||
| rs4951059 | chr1 | 204258018 | PHACTR1 | 221692 | 0.07 | trans | ||
| rs17014160 | chr1 | 206855799 | PHACTR1 | 221692 | 0.01 | trans | ||
| snp_a-2090942 | 0 | PHACTR1 | 221692 | 0.08 | trans | |||
| rs17014165 | chr1 | 206856485 | PHACTR1 | 221692 | 0.08 | trans | ||
| rs6733654 | chr2 | 1963270 | PHACTR1 | 221692 | 0.02 | trans | ||
| rs2712089 | chr2 | 24080273 | PHACTR1 | 221692 | 0.04 | trans | ||
| rs6544404 | chr2 | 41502544 | PHACTR1 | 221692 | 0.19 | trans | ||
| rs17029291 | chr3 | 32402138 | PHACTR1 | 221692 | 6.006E-5 | trans | ||
| rs1553189 | chr3 | 112939422 | PHACTR1 | 221692 | 0.19 | trans | ||
| rs1807299 | chr3 | 134470799 | PHACTR1 | 221692 | 0.03 | trans | ||
| rs830202 | chr3 | 165654796 | PHACTR1 | 221692 | 0.04 | trans | ||
| rs2339917 | chr3 | 179669447 | PHACTR1 | 221692 | 0.04 | trans | ||
| rs6813178 | chr4 | 71575238 | PHACTR1 | 221692 | 0.01 | trans | ||
| rs6843749 | chr4 | 84847249 | PHACTR1 | 221692 | 0.07 | trans | ||
| rs16901958 | chr5 | 44776555 | PHACTR1 | 221692 | 0.15 | trans | ||
| rs750544 | chr5 | 51749054 | PHACTR1 | 221692 | 0.02 | trans | ||
| rs17146240 | chr5 | 119522933 | PHACTR1 | 221692 | 0.01 | trans | ||
| rs17146261 | chr5 | 119534252 | PHACTR1 | 221692 | 0.01 | trans | ||
| rs2807510 | chr6 | 73090536 | PHACTR1 | 221692 | 0.08 | trans | ||
| rs542717 | chr6 | 75934072 | PHACTR1 | 221692 | 0.13 | trans | ||
| rs4617108 | chr7 | 49423831 | PHACTR1 | 221692 | 0.11 | trans | ||
| rs3779536 | chr7 | 127233976 | PHACTR1 | 221692 | 0.07 | trans | ||
| rs3105341 | chr10 | 58849852 | PHACTR1 | 221692 | 0.18 | trans | ||
| rs1717532 | chr10 | 59017025 | PHACTR1 | 221692 | 0.13 | trans | ||
| rs12569493 | chr10 | 74886042 | PHACTR1 | 221692 | 0.12 | trans | ||
| rs16930362 | chr10 | 74890320 | PHACTR1 | 221692 | 0.04 | trans | ||
| rs7092445 | chr10 | 74921991 | PHACTR1 | 221692 | 0.09 | trans | ||
| rs11000607 | chr10 | 75089777 | PHACTR1 | 221692 | 0.13 | trans | ||
| rs12571454 | chr10 | 75208954 | PHACTR1 | 221692 | 0.08 | trans | ||
| rs1700369 | chr12 | 127842232 | PHACTR1 | 221692 | 0.02 | trans | ||
| rs1324669 | chr13 | 107872446 | PHACTR1 | 221692 | 9.699E-4 | trans | ||
| rs3742829 | chr14 | 73489267 | PHACTR1 | 221692 | 0.15 | trans | ||
| rs8055506 | chr16 | 13661723 | PHACTR1 | 221692 | 0.18 | trans | ||
| rs16962446 | chr16 | 13694936 | PHACTR1 | 221692 | 0.04 | trans | ||
| rs2239335 | chr16 | 22897794 | PHACTR1 | 221692 | 0.06 | trans | ||
| rs9937567 | chr16 | 66364752 | PHACTR1 | 221692 | 0.08 | trans | ||
| rs6098023 | chr20 | 53113740 | PHACTR1 | 221692 | 0 | trans | ||
| rs6015314 | chr20 | 57167224 | PHACTR1 | 221692 | 0.01 | trans | ||
| rs12157904 | chr22 | 37982011 | PHACTR1 | 221692 | 0.13 | trans | ||
| rs17145698 | chrX | 40218345 | PHACTR1 | 221692 | 0.01 | trans |
Section II. Transcriptome annotation
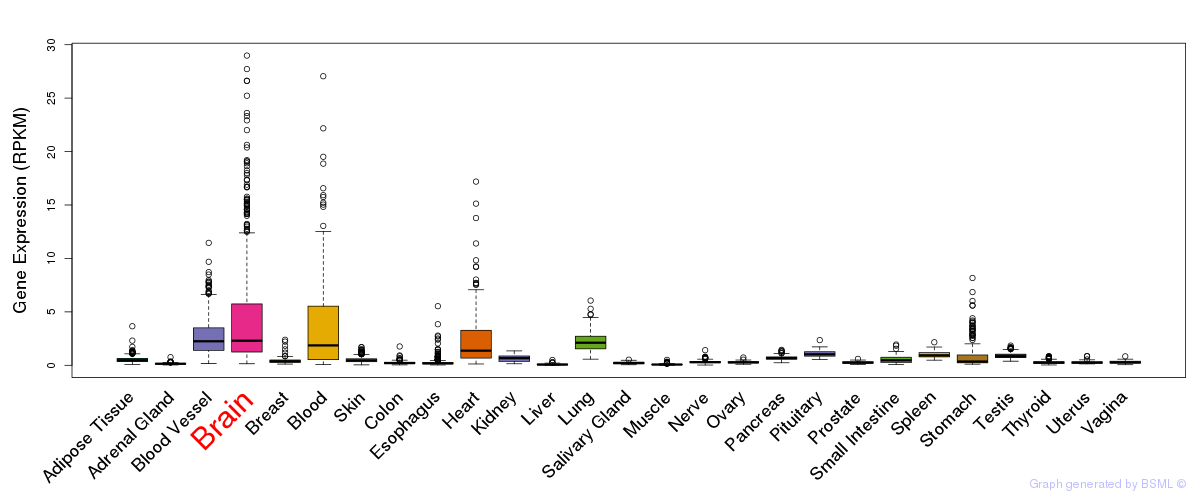
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0003779 | actin binding | IEA | - | |
| GO:0004864 | protein phosphatase inhibitor activity | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0045202 | synapse | IEA | neuron, Synap, Neurotransmitter, Glial (GO term level: 2) | - |
| GO:0005737 | cytoplasm | IEA | - | |
| GO:0030054 | cell junction | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| ONKEN UVEAL MELANOMA UP | 783 | 507 | All SZGR 2.0 genes in this pathway |
| JAATINEN HEMATOPOIETIC STEM CELL UP | 316 | 190 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA UP | 1821 | 933 | All SZGR 2.0 genes in this pathway |
| PUJANA ATM PCC NETWORK | 1442 | 892 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 48HR DN | 428 | 306 | All SZGR 2.0 genes in this pathway |
| NIKOLSKY BREAST CANCER 6P24 P22 AMPLICON | 21 | 17 | All SZGR 2.0 genes in this pathway |
| BASSO CD40 SIGNALING UP | 101 | 76 | All SZGR 2.0 genes in this pathway |
| DAZARD RESPONSE TO UV NHEK UP | 244 | 151 | All SZGR 2.0 genes in this pathway |
| DAZARD UV RESPONSE CLUSTER G1 | 67 | 41 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 6HR DN | 911 | 527 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 24HR DN | 1011 | 592 | All SZGR 2.0 genes in this pathway |
| KONDO PROSTATE CANCER WITH H3K27ME3 | 196 | 93 | All SZGR 2.0 genes in this pathway |
| ZHANG GATA6 TARGETS DN | 64 | 46 | All SZGR 2.0 genes in this pathway |
| RAY TUMORIGENESIS BY ERBB2 CDC25A DN | 159 | 105 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO CSF2RB AND IL4 DN | 315 | 201 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO HGF DN | 235 | 144 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO HGF VS CSF2RB AND IL4 UP | 408 | 276 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN MEF ICP WITH H3K27ME3 | 206 | 108 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN MEF HCP WITH H3K27ME3 | 590 | 403 | All SZGR 2.0 genes in this pathway |
| WIERENGA STAT5A TARGETS UP | 217 | 131 | All SZGR 2.0 genes in this pathway |
| WIERENGA STAT5A TARGETS GROUP1 | 136 | 76 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE VIA TP53 GROUP D | 280 | 158 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-335 | 12 | 18 | 1A | hsa-miR-335brain | UCAAGAGCAAUAACGAAAAAUGU |
| miR-34b | 106 | 112 | m8 | hsa-miR-34b | UAGGCAGUGUCAUUAGCUGAUUG |
| miR-496 | 91 | 97 | 1A | hsa-miR-496 | AUUACAUGGCCAAUCUC |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.