Gene Page: FHL3
Summary ?
| GeneID | 2275 |
| Symbol | FHL3 |
| Synonyms | SLIM2 |
| Description | four and a half LIM domains 3 |
| Reference | MIM:602790|HGNC:HGNC:3704|Ensembl:ENSG00000183386|HPRD:09101|Vega:OTTHUMG00000004434 |
| Gene type | protein-coding |
| Map location | 1p34 |
| Pascal p-value | 0.061 |
| Sherlock p-value | 0.332 |
| Fetal beta | 0.264 |
| DMG | 1 (# studies) |
| eGene | Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Nishioka_2013 | Genome-wide DNA methylation analysis | The authors investigated the methylation profiles of DNA in peripheral blood cells from 18 patients with first-episode schizophrenia (FESZ) and from 15 normal controls. | 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg27018070 | 1 | 38470578 | FHL3 | -0.024 | 0.78 | DMG:Nishioka_2013 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs17275845 | chr2 | 213836589 | FHL3 | 2275 | 0.19 | trans |
Section II. Transcriptome annotation
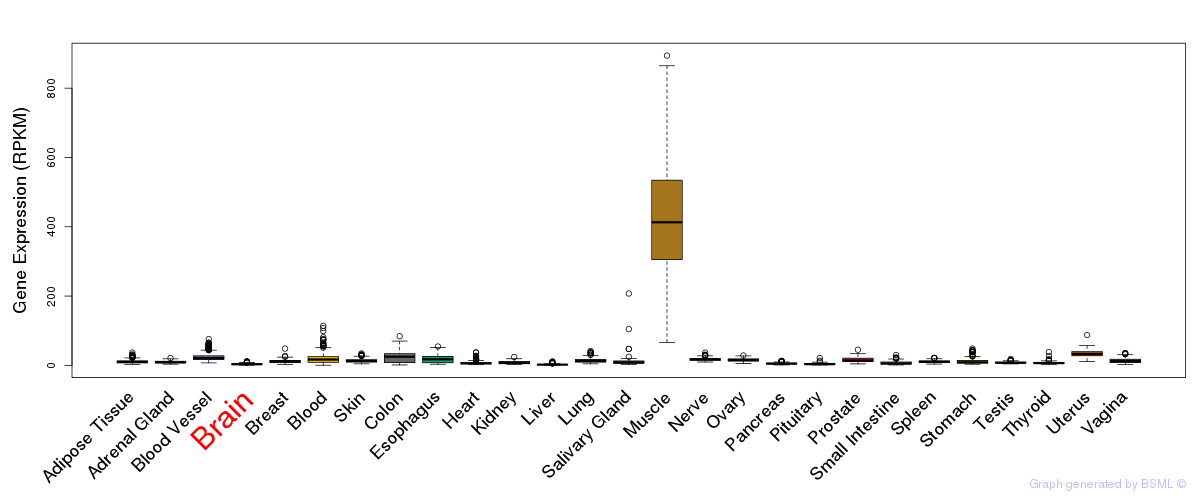
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ADAMTSL4 | TSRC1 | ADAMTS-like 4 | Two-hybrid | BioGRID | 16189514 |
| CDC25B | - | cell division cycle 25 homolog B (S. pombe) | - | HPRD | 12681290 |
| CDC42EP1 | BORG5 | CEP1 | MGC15316 | MSE55 | CDC42 effector protein (Rho GTPase binding) 1 | Two-hybrid | BioGRID | 16189514 |
| CREB1 | CREB | MGC9284 | cAMP responsive element binding protein 1 | Affinity Capture-Western Two-hybrid | BioGRID | 11046156 |
| CTBP2 | - | C-terminal binding protein 2 | - | HPRD,BioGRID | 12556451 |
| FHL2 | AAG11 | DRAL | SLIM3 | four and a half LIM domains 2 | - | HPRD,BioGRID | 11135358 |
| FHL3 | MGC19547 | MGC23614 | MGC8696 | SLIM2 | four and a half LIM domains 3 | Two-hybrid | BioGRID | 11046156 |
| FHL3 | MGC19547 | MGC23614 | MGC8696 | SLIM2 | four and a half LIM domains 3 | - | HPRD | 15117962 |
| FHL5 | ACT | FLJ33049 | KIAA0776 | RP3-393D12.2 | dJ393D12.2 | four and a half LIM domains 5 | Two-hybrid | BioGRID | 11046156 |
| ITGA7 | FLJ25220 | integrin, alpha 7 | Affinity Capture-Western Co-localization Two-hybrid | BioGRID | 15117962 |
| JTV1 | AIMP2 | P38 | PRO0992 | JTV1 gene | Two-hybrid | BioGRID | 16189514 |
| KLF3 | BKLF | MGC48279 | Kruppel-like factor 3 (basic) | Affinity Capture-Western Reconstituted Complex Two-hybrid | BioGRID | 12556451 |
| KLF8 | BKLF3 | DKFZp686O08126 | DXS741 | MGC138314 | ZNF741 | Kruppel-like factor 8 | - | HPRD | 12556451 |
| LASP1 | Lasp-1 | MLN50 | LIM and SH3 protein 1 | Two-hybrid | BioGRID | 16189514 |
| LNX1 | LNX | MPDZ | PDZRN2 | ligand of numb-protein X 1 | Two-hybrid | BioGRID | 16189514 |
| MAPK1 | ERK | ERK2 | ERT1 | MAPK2 | P42MAPK | PRKM1 | PRKM2 | p38 | p40 | p41 | p41mapk | mitogen-activated protein kinase 1 | - | HPRD | 14729955 |
| MED15 | ARC105 | CAG7A | CTG7A | DKFZp686A2214 | DKFZp762B1216 | FLJ42282 | FLJ42935 | PCQAP | TIG-1 | TIG1 | TNRC7 | mediator complex subunit 15 | Two-hybrid | BioGRID | 16189514 |
| PAK7 | KIAA1264 | MGC26232 | PAK5 | p21 protein (Cdc42/Rac)-activated kinase 7 | Two-hybrid | BioGRID | 16189514 |
| PHC2 | EDR2 | HPH2 | MGC163502 | PH2 | polyhomeotic homolog 2 (Drosophila) | Two-hybrid | BioGRID | 16189514 |
| RBM42 | MGC10433 | RNA binding motif protein 42 | Two-hybrid | BioGRID | 16189514 |
| RFX6 | MGC33442 | RFXDC1 | dJ955L16.1 | regulatory factor X, 6 | Two-hybrid | BioGRID | 16189514 |
| SRF | MCM1 | serum response factor (c-fos serum response element-binding transcription factor) | SRF interacts with FHL3. This interaction was modeled on a demonstrated interaction between human SRF and FHL3 from an unspecified species. | BIND | 15610731 |
| TRIP6 | MGC10556 | MGC10558 | MGC29959 | MGC3837 | MGC4423 | OIP1 | ZRP-1 | thyroid hormone receptor interactor 6 | Two-hybrid | BioGRID | 16189514 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| PARENT MTOR SIGNALING DN | 46 | 29 | All SZGR 2.0 genes in this pathway |
| AMIT EGF RESPONSE 240 HELA | 60 | 43 | All SZGR 2.0 genes in this pathway |
| WESTON VEGFA TARGETS 6HR | 59 | 38 | All SZGR 2.0 genes in this pathway |
| WESTON VEGFA TARGETS | 108 | 71 | All SZGR 2.0 genes in this pathway |
| DOUGLAS BMI1 TARGETS UP | 566 | 371 | All SZGR 2.0 genes in this pathway |
| MEISSNER BRAIN HCP WITH H3K4ME3 AND H3K27ME3 | 1069 | 729 | All SZGR 2.0 genes in this pathway |
| ROME INSULIN TARGETS IN MUSCLE UP | 442 | 263 | All SZGR 2.0 genes in this pathway |
| HOSHIDA LIVER CANCER SUBCLASS S1 | 237 | 159 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE VIA TP53 GROUP B | 549 | 316 | All SZGR 2.0 genes in this pathway |
| KATSANOU ELAVL1 TARGETS DN | 148 | 88 | All SZGR 2.0 genes in this pathway |
| WAKABAYASHI ADIPOGENESIS PPARG BOUND 8D | 658 | 397 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 2ND EGF PULSE ONLY | 1725 | 838 | All SZGR 2.0 genes in this pathway |