Gene Page: XRN2
Summary ?
| GeneID | 22803 |
| Symbol | XRN2 |
| Synonyms | - |
| Description | 5'-3' exoribonuclease 2 |
| Reference | MIM:608851|HGNC:HGNC:12836|Ensembl:ENSG00000088930|HPRD:10309|Vega:OTTHUMG00000032025 |
| Gene type | protein-coding |
| Map location | 20p11.2-p11.1 |
| Pascal p-value | 3.392E-4 |
| Sherlock p-value | 0.193 |
| Fetal beta | 1.751 |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 1.9018 |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
General gene expression (GTEx)
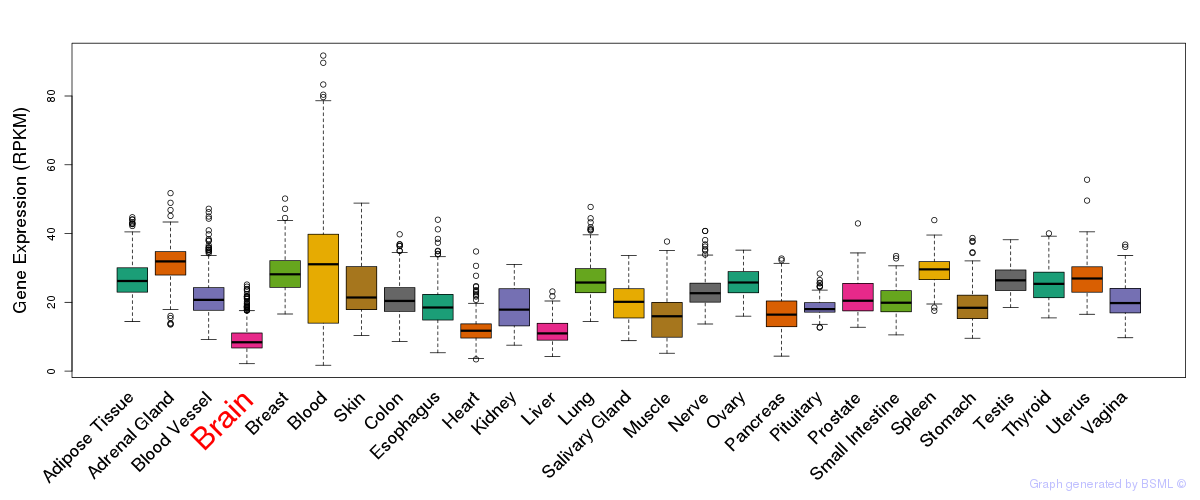
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| KIF13A | 0.84 | 0.84 |
| CC2D1B | 0.81 | 0.84 |
| PHLDB1 | 0.81 | 0.83 |
| FRYL | 0.80 | 0.81 |
| KIAA0562 | 0.79 | 0.82 |
| ZFYVE16 | 0.79 | 0.79 |
| RNF213 | 0.79 | 0.81 |
| ATP11A | 0.79 | 0.80 |
| WWC3 | 0.79 | 0.82 |
| XPC | 0.79 | 0.81 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| UQCR | -0.50 | -0.60 |
| AC044839.2 | -0.49 | -0.45 |
| C1orf61 | -0.49 | -0.45 |
| NDUFB2 | -0.49 | -0.50 |
| C1orf54 | -0.49 | -0.48 |
| TIMM8B | -0.49 | -0.51 |
| UQCRQ | -0.48 | -0.54 |
| ENHO | -0.48 | -0.48 |
| IL32 | -0.48 | -0.50 |
| SERF2 | -0.48 | -0.50 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0003676 | nucleic acid binding | IEA | - | |
| GO:0005515 | protein binding | IPI | 15231747 | |
| GO:0004534 | 5'-3' exoribonuclease activity | TAS | 10409438 | |
| GO:0016787 | hydrolase activity | IEA | - | |
| GO:0008270 | zinc ion binding | IEA | - | |
| GO:0046872 | metal ion binding | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0000738 | DNA catabolic process, exonucleolytic | IDA | 15565158 | |
| GO:0006353 | transcription termination | IEA | - | |
| GO:0006355 | regulation of transcription, DNA-dependent | IEA | - | |
| GO:0006397 | mRNA processing | IEA | - | |
| GO:0006350 | transcription | IEA | - | |
| GO:0006401 | RNA catabolic process | TAS | 10409438 | |
| GO:0016049 | cell growth | ISS | - | |
| GO:0007283 | spermatogenesis | IEP | 10409438 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005622 | intracellular | IEA | - | |
| GO:0005634 | nucleus | TAS | 10409438 | |
| GO:0005730 | nucleolus | IDA | 12429849 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| COMT | - | catechol-O-methyltransferase | - | HPRD,BioGRID | 15231747 |
| DOM3Z | DOM3L | NG6 | dom-3 homolog Z (C. elegans) | - | HPRD,BioGRID | 15231747 |
| EEF1A1 | CCS-3 | CCS3 | EEF-1 | EEF1A | EF-Tu | EF1A | FLJ25721 | GRAF-1EF | HNGC:16303 | LENG7 | MGC102687 | MGC131894 | MGC16224 | PTI1 | eEF1A-1 | eukaryotic translation elongation factor 1 alpha 1 | - | HPRD,BioGRID | 15231747 |
| EIF5A | A | EIF-5A | EIF5A1 | MGC104255 | MGC99547 | uORF | eukaryotic translation initiation factor 5A | - | HPRD,BioGRID | 15231747 |
| EXOSC10 | PM-Scl | PM/Scl-100 | PMSCL | PMSCL2 | RRP6 | Rrp6p | p2 | p3 | p4 | exosome component 10 | - | HPRD,BioGRID | 15231747 |
| LSM3 | SMX4 | USS2 | YLR438C | LSM3 homolog, U6 small nuclear RNA associated (S. cerevisiae) | - | HPRD,BioGRID | 15231747 |
| MOCS3 | MGC9252 | UBA4 | dJ914P20.3 | molybdenum cofactor synthesis 3 | - | HPRD,BioGRID | 15231747 |
| PSMA3 | HC8 | MGC12306 | MGC32631 | PSC3 | proteasome (prosome, macropain) subunit, alpha type, 3 | - | HPRD,BioGRID | 15231747 |
| TARDBP | ALS10 | TDP-43 | TAR DNA binding protein | - | HPRD,BioGRID | 15231747 |
| TOLLIP | FLJ33531 | IL-1RAcPIP | toll interacting protein | Two-hybrid | BioGRID | 15231747 |
| UPF2 | DKFZp434D222 | HUPF2 | KIAA1408 | MGC138834 | MGC138835 | RENT2 | smg-3 | UPF2 regulator of nonsense transcripts homolog (yeast) | - | HPRD,BioGRID | 15231747 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG RNA DEGRADATION | 59 | 37 | All SZGR 2.0 genes in this pathway |
| REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | 27 | 15 | All SZGR 2.0 genes in this pathway |
| REACTOME PROTEIN FOLDING | 53 | 30 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF PROTEINS | 518 | 242 | All SZGR 2.0 genes in this pathway |
| FULCHER INFLAMMATORY RESPONSE LECTIN VS LPS DN | 463 | 290 | All SZGR 2.0 genes in this pathway |
| KINSEY TARGETS OF EWSR1 FLII FUSION UP | 1278 | 748 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA ANAPLASTIC UP | 722 | 443 | All SZGR 2.0 genes in this pathway |
| GOTZMANN EPITHELIAL TO MESENCHYMAL TRANSITION DN | 206 | 136 | All SZGR 2.0 genes in this pathway |
| ROZANOV MMP14 TARGETS UP | 266 | 171 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 TARGETS UP | 673 | 430 | All SZGR 2.0 genes in this pathway |
| MARTINEZ TP53 TARGETS UP | 602 | 364 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 AND TP53 TARGETS UP | 601 | 369 | All SZGR 2.0 genes in this pathway |
| GRADE COLON AND RECTAL CANCER UP | 285 | 167 | All SZGR 2.0 genes in this pathway |
| CHIANG LIVER CANCER SUBCLASS UNANNOTATED DN | 193 | 112 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS DN | 1972 | 1213 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE LATE | 1137 | 655 | All SZGR 2.0 genes in this pathway |
| FEVR CTNNB1 TARGETS DN | 553 | 343 | All SZGR 2.0 genes in this pathway |
| RAO BOUND BY SALL4 | 227 | 149 | All SZGR 2.0 genes in this pathway |
| PECE MAMMARY STEM CELL DN | 146 | 88 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-203.1 | 411 | 418 | 1A,m8 | hsa-miR-203 | UGAAAUGUUUAGGACCACUAG |
| miR-539 | 68 | 74 | 1A | hsa-miR-539 | GGAGAAAUUAUCCUUGGUGUGU |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.