Gene Page: RAB11FIP2
Summary ?
| GeneID | 22841 |
| Symbol | RAB11FIP2 |
| Synonyms | Rab11-FIP2|nRip11 |
| Description | RAB11 family interacting protein 2 (class I) |
| Reference | MIM:608599|HGNC:HGNC:29152|Ensembl:ENSG00000107560|HPRD:10548|Vega:OTTHUMG00000019128 |
| Gene type | protein-coding |
| Map location | 10q26.11 |
| Pascal p-value | 0.414 |
| Sherlock p-value | 0.547 |
| Fetal beta | -0.352 |
| DMG | 1 (# studies) |
| Support | G2Cdb.human_BAYES-COLLINS-MOUSE-PSD-CONSENSUS G2Cdb.humanPSD G2Cdb.humanPSP |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg02291245 | 10 | 119780037 | RAB11FIP2 | 2.269E-4 | -0.299 | 0.036 | DMG:Wockner_2014 |
Section II. Transcriptome annotation
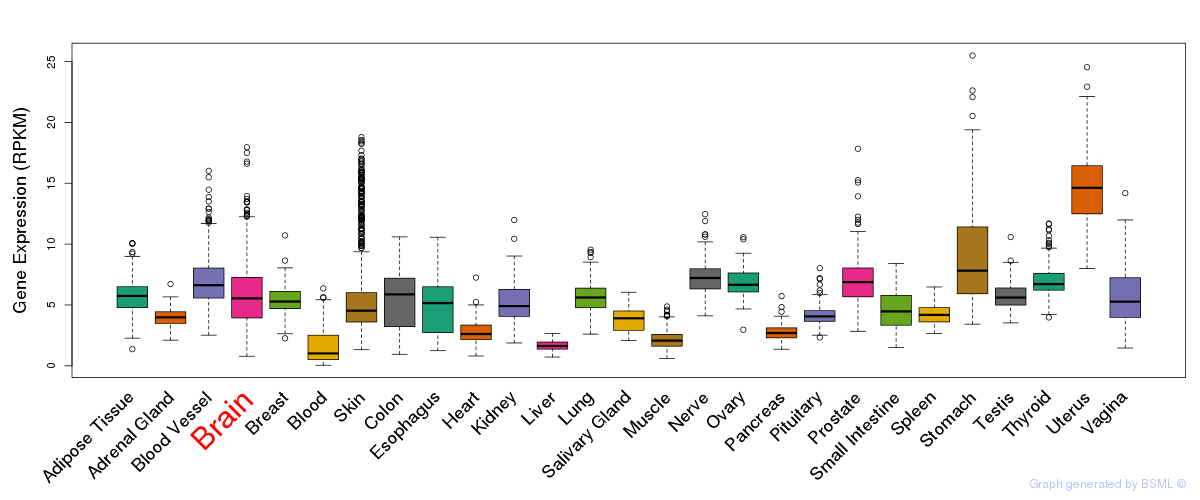
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| GRIN2A | 0.82 | 0.74 |
| KCNB1 | 0.81 | 0.65 |
| TRHDE | 0.81 | 0.69 |
| PLCB1 | 0.80 | 0.59 |
| KLF9 | 0.80 | 0.70 |
| CREG2 | 0.79 | 0.66 |
| TMEM132D | 0.79 | 0.74 |
| GCC2 | 0.79 | 0.74 |
| ARHGAP26 | 0.79 | 0.72 |
| CMTM4 | 0.79 | 0.68 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| BCL7C | -0.46 | -0.62 |
| AC135586.1 | -0.41 | -0.53 |
| TRAF4 | -0.41 | -0.57 |
| UBE2L6 | -0.41 | -0.46 |
| RPL18 | -0.40 | -0.62 |
| RPL35 | -0.40 | -0.57 |
| FAU | -0.40 | -0.55 |
| RPLP1 | -0.40 | -0.56 |
| RPL36 | -0.40 | -0.59 |
| RPS18 | -0.40 | -0.58 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| AP2A1 | ADTAA | AP2-ALPHA | CLAPA1 | adaptor-related protein complex 2, alpha 1 subunit | - | HPRD,BioGRID | 12364336 |
| EPS15 | AF-1P | AF1P | MLLT5 | epidermal growth factor receptor pathway substrate 15 | Affinity Capture-Western | BioGRID | 12364336 |
| IL8RB | CD182 | CDw128b | CMKAR2 | CXCR2 | IL8R2 | IL8RA | interleukin 8 receptor, beta | - | HPRD,BioGRID | 15004234 |
| MYO5B | KIAA1119 | myosin VB | - | HPRD,BioGRID | 11495908 |12393859 |
| RAB11A | MGC1490 | YL8 | RAB11A, member RAS oncogene family | - | HPRD | 15304524 |
| RAB11A | MGC1490 | YL8 | RAB11A, member RAS oncogene family | - | HPRD,BioGRID | 11495908 |11994279 |
| RAB11B | H-YPT3 | MGC133246 | RAB11B, member RAS oncogene family | - | HPRD,BioGRID | 11495908 |
| RAB11FIP1 | DKFZp686E2214 | FLJ22524 | FLJ22622 | MGC78448 | NOEL1A | RCP | rab11-FIP1 | RAB11 family interacting protein 1 (class I) | Affinity Capture-Western | BioGRID | 12364336 |
| RAB11FIP2 | KIAA0941 | Rab11-FIP2 | nRip11 | RAB11 family interacting protein 2 (class I) | Affinity Capture-Western Two-hybrid | BioGRID | 11994279 |12364336 |
| RAB11FIP4 | FLJ00131 | KIAA1821 | MGC11316 | MGC126566 | RAB11-FIP4 | RAB11 family interacting protein 4 (class II) | - | HPRD,BioGRID | 12470645 |
| RAB11FIP5 | DKFZp434H018 | GAF1 | KIAA0857 | RIP11 | pp75 | RAB11 family interacting protein 5 (class I) | Affinity Capture-Western | BioGRID | 12364336 |
| RAB25 | CATX-8 | RAB25, member RAS oncogene family | - | HPRD,BioGRID | 11481332 |
| RAB25 | CATX-8 | RAB25, member RAS oncogene family | - | HPRD | 11495908 |
| REPS1 | RALBP1 | RALBP1 associated Eps domain containing 1 | Affinity Capture-Western | BioGRID | 12364336 |
| YWHAG | 14-3-3GAMMA | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, gamma polypeptide | - | HPRD | 15324660 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG ENDOCYTOSIS | 183 | 132 | All SZGR 2.0 genes in this pathway |
| REACTOME TRANSMEMBRANE TRANSPORT OF SMALL MOLECULES | 413 | 270 | All SZGR 2.0 genes in this pathway |
| REACTOME AQUAPORIN MEDIATED TRANSPORT | 51 | 34 | All SZGR 2.0 genes in this pathway |
| REACTOME REGULATION OF WATER BALANCE BY RENAL AQUAPORINS | 44 | 30 | All SZGR 2.0 genes in this pathway |
| PUIFFE INVASION INHIBITED BY ASCITES DN | 145 | 91 | All SZGR 2.0 genes in this pathway |
| WAMUNYOKOLI OVARIAN CANCER LMP DN | 199 | 124 | All SZGR 2.0 genes in this pathway |
| LASTOWSKA NEUROBLASTOMA COPY NUMBER DN | 800 | 473 | All SZGR 2.0 genes in this pathway |
| GEORGES TARGETS OF MIR192 AND MIR215 | 893 | 528 | All SZGR 2.0 genes in this pathway |
| ZHANG RESPONSE TO IKK INHIBITOR AND TNF DN | 103 | 64 | All SZGR 2.0 genes in this pathway |
| DEBIASI APOPTOSIS BY REOVIRUS INFECTION UP | 314 | 201 | All SZGR 2.0 genes in this pathway |
| CHANG CORE SERUM RESPONSE DN | 209 | 137 | All SZGR 2.0 genes in this pathway |
| HAN SATB1 TARGETS DN | 442 | 275 | All SZGR 2.0 genes in this pathway |
| CHIANG LIVER CANCER SUBCLASS CTNNB1 UP | 176 | 110 | All SZGR 2.0 genes in this pathway |
| LU EZH2 TARGETS DN | 414 | 237 | All SZGR 2.0 genes in this pathway |
| GABRIELY MIR21 TARGETS | 289 | 187 | All SZGR 2.0 genes in this pathway |
| LINSLEY MIR16 TARGETS | 206 | 127 | All SZGR 2.0 genes in this pathway |
| GOBERT OLIGODENDROCYTE DIFFERENTIATION DN | 1080 | 713 | All SZGR 2.0 genes in this pathway |
| PHONG TNF RESPONSE NOT VIA P38 | 337 | 236 | All SZGR 2.0 genes in this pathway |