Gene Page: CHSY1
Summary ?
| GeneID | 22856 |
| Symbol | CHSY1 |
| Synonyms | CHSY|CSS1|ChSy-1|TPBS |
| Description | chondroitin sulfate synthase 1 |
| Reference | MIM:608183|HGNC:HGNC:17198|Ensembl:ENSG00000131873|HPRD:10493|Vega:OTTHUMG00000149873 |
| Gene type | protein-coding |
| Map location | 15q26.3 |
| Pascal p-value | 0.328 |
| Sherlock p-value | 0.136 |
| TADA p-value | 0.014 |
| Fetal beta | 0.795 |
| DMG | 1 (# studies) |
| eGene | Cerebellum Cortex Myers' cis & trans |
| Support | CompositeSet Ascano FMRP targets |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 1 |
| DNM:Fromer_2014 | Whole Exome Sequencing analysis | This study reported a WES study of 623 schizophrenia trios, reporting DNMs using genomic DNA. |
Section I. Genetics and epigenetics annotation
 DNM table
DNM table
| Gene | Chromosome | Position | Ref | Alt | Transcript | AA change | Mutation type | Sift | CG46 | Trait | Study |
|---|---|---|---|---|---|---|---|---|---|---|---|
| CHSY1 | chr15 | 101718038 | T | C | NM_014918 | p.655Y>C | missense | Schizophrenia | DNM:Fromer_2014 |
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg27425263 | 15 | 101792104 | CHSY1 | 1.56E-8 | -0.013 | 5.84E-6 | DMG:Jaffe_2016 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs6047026 | chr20 | 20730673 | CHSY1 | 22856 | 0.14 | trans | ||
| rs6075703 | chr20 | 20736750 | CHSY1 | 22856 | 0.14 | trans |
Section II. Transcriptome annotation
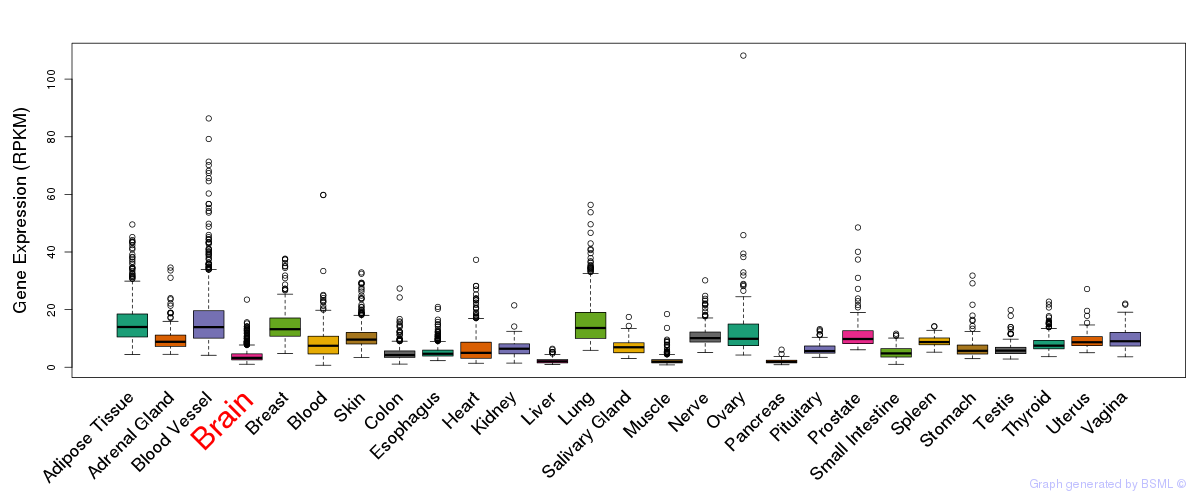
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| KIF5A | 0.89 | 0.89 |
| PPP2R2C | 0.80 | 0.87 |
| COBL | 0.80 | 0.81 |
| LRRK2 | 0.80 | 0.85 |
| RIMS2 | 0.79 | 0.82 |
| TOM1L2 | 0.79 | 0.80 |
| IQSEC1 | 0.79 | 0.78 |
| PACSIN1 | 0.79 | 0.78 |
| IQSEC3 | 0.79 | 0.82 |
| SUSD5 | 0.79 | 0.82 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| BCL7C | -0.54 | -0.61 |
| RPL18 | -0.53 | -0.62 |
| RPL27 | -0.52 | -0.59 |
| RPS16P1 | -0.52 | -0.64 |
| RPL35 | -0.52 | -0.60 |
| RPL28 | -0.52 | -0.61 |
| RPS15A | -0.52 | -0.62 |
| UXT | -0.51 | -0.55 |
| RPS19P3 | -0.51 | -0.61 |
| RPS3P3 | -0.51 | -0.56 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG GLYCOSAMINOGLYCAN BIOSYNTHESIS CHONDROITIN SULFATE | 22 | 16 | All SZGR 2.0 genes in this pathway |
| REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | 21 | 15 | All SZGR 2.0 genes in this pathway |
| REACTOME CHONDROITIN SULFATE DERMATAN SULFATE METABOLISM | 49 | 33 | All SZGR 2.0 genes in this pathway |
| REACTOME GLYCOSAMINOGLYCAN METABOLISM | 111 | 69 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF CARBOHYDRATES | 247 | 154 | All SZGR 2.0 genes in this pathway |
| LIU TARGETS OF VMYB VS CMYB DN | 43 | 30 | All SZGR 2.0 genes in this pathway |
| CHARAFE BREAST CANCER LUMINAL VS MESENCHYMAL DN | 460 | 312 | All SZGR 2.0 genes in this pathway |
| LINDGREN BLADDER CANCER CLUSTER 2B | 392 | 251 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 DN | 855 | 609 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 COMMON DN | 483 | 336 | All SZGR 2.0 genes in this pathway |
| NUYTTEN NIPP1 TARGETS UP | 769 | 437 | All SZGR 2.0 genes in this pathway |
| NUYTTEN EZH2 TARGETS UP | 1037 | 673 | All SZGR 2.0 genes in this pathway |
| NIKOLSKY BREAST CANCER 15Q26 AMPLICON | 22 | 19 | All SZGR 2.0 genes in this pathway |
| ROZANOV MMP14 TARGETS SUBSET | 33 | 20 | All SZGR 2.0 genes in this pathway |
| ROZANOV MMP14 TARGETS UP | 266 | 171 | All SZGR 2.0 genes in this pathway |
| MANALO HYPOXIA UP | 207 | 145 | All SZGR 2.0 genes in this pathway |
| GENTILE UV RESPONSE CLUSTER D4 | 55 | 37 | All SZGR 2.0 genes in this pathway |
| LU AGING BRAIN UP | 262 | 186 | All SZGR 2.0 genes in this pathway |
| GENTILE UV HIGH DOSE DN | 312 | 203 | All SZGR 2.0 genes in this pathway |
| IWANAGA CARCINOGENESIS BY KRAS PTEN DN | 353 | 226 | All SZGR 2.0 genes in this pathway |
| MCCABE BOUND BY HOXC6 | 469 | 239 | All SZGR 2.0 genes in this pathway |
| CHEN METABOLIC SYNDROM NETWORK | 1210 | 725 | All SZGR 2.0 genes in this pathway |
| MILI PSEUDOPODIA HAPTOTAXIS DN | 668 | 419 | All SZGR 2.0 genes in this pathway |
| HOSHIDA LIVER CANCER SURVIVAL UP | 73 | 49 | All SZGR 2.0 genes in this pathway |
| LI INDUCED T TO NATURAL KILLER UP | 307 | 182 | All SZGR 2.0 genes in this pathway |
| BHAT ESR1 TARGETS NOT VIA AKT1 UP | 211 | 131 | All SZGR 2.0 genes in this pathway |
| GOBERT OLIGODENDROCYTE DIFFERENTIATION UP | 570 | 339 | All SZGR 2.0 genes in this pathway |
| WAKABAYASHI ADIPOGENESIS PPARG BOUND 8D | 658 | 397 | All SZGR 2.0 genes in this pathway |
| KRIEG HYPOXIA NOT VIA KDM3A | 770 | 480 | All SZGR 2.0 genes in this pathway |
| FORTSCHEGGER PHF8 TARGETS DN | 784 | 464 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 1ST EGF PULSE ONLY | 1839 | 928 | All SZGR 2.0 genes in this pathway |