Gene Page: CNKSR2
Summary ?
| GeneID | 22866 |
| Symbol | CNKSR2 |
| Synonyms | CNK2|KSR2|MAGUIN |
| Description | connector enhancer of kinase suppressor of Ras 2 |
| Reference | MIM:300724|HGNC:HGNC:19701|Ensembl:ENSG00000149970|HPRD:06473|Vega:OTTHUMG00000021233 |
| Gene type | protein-coding |
| Map location | Xp22.12 |
| Fetal beta | -0.866 |
| Support | PROTEIN CLUSTERING G2Cdb.human_BAYES-COLLINS-MOUSE-PSD-CONSENSUS G2Cdb.humanPSD G2Cdb.humanPSP Potential synaptic genes |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWAScat | Genome-wide Association Studies | This data set includes 560 SNPs associated with schizophrenia. A total of 486 genes were mapped to these SNPs within 50kb. | |
| CV:PGC128 | Genome-wide Association Study | A multi-stage schizophrenia GWAS of up to 36,989 cases and 113,075 controls. Reported by the Schizophrenia Working Group of PGC. 128 independent associations spanning 108 loci | |
| CV:PGCnp | Genome-wide Association Study | GWAS |
Section I. Genetics and epigenetics annotation
 CV:PGC128
CV:PGC128
| SNP ID | Chromosome | Position | Allele | P | Function | Gene | Up/Down Distance |
|---|---|---|---|---|---|---|---|
| rs1378559 | chrX | 21380266 | TC | 1.679E-11 | intergenic | RPS6KA3,CNKSR2 | dist=1095516;dist=12270 |
Section II. Transcriptome annotation
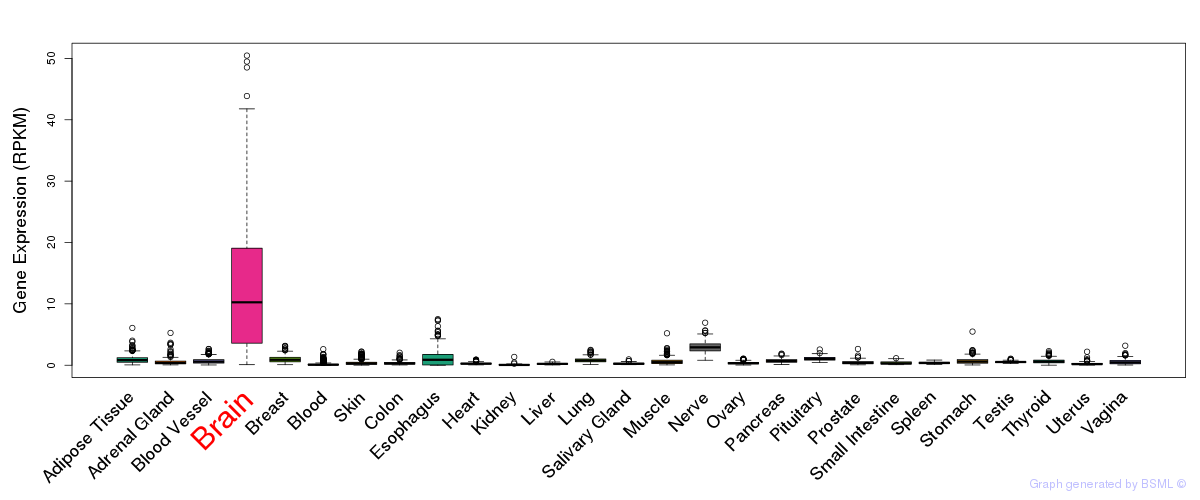
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| KIAA0494 | 0.89 | 0.87 |
| MOSPD2 | 0.88 | 0.84 |
| AP3B1 | 0.87 | 0.86 |
| STXBP3 | 0.87 | 0.82 |
| MAGT1 | 0.87 | 0.85 |
| MTMR10 | 0.86 | 0.87 |
| GAB1 | 0.85 | 0.86 |
| PXK | 0.85 | 0.79 |
| MPP5 | 0.85 | 0.85 |
| BBX | 0.85 | 0.85 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| TBC1D10A | -0.47 | -0.45 |
| C1orf86 | -0.45 | -0.48 |
| WDR86 | -0.45 | -0.47 |
| EMID1 | -0.44 | -0.47 |
| NANOS3 | -0.42 | -0.54 |
| COMTD1 | -0.41 | -0.41 |
| CCDC28B | -0.40 | -0.47 |
| C6orf129 | -0.40 | -0.47 |
| PHPT1 | -0.40 | -0.46 |
| AC174470.1 | -0.40 | -0.41 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| ZHOU INFLAMMATORY RESPONSE FIMA DN | 284 | 156 | All SZGR 2.0 genes in this pathway |
| PUJANA ATM PCC NETWORK | 1442 | 892 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE DN | 1237 | 837 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS UP | 1305 | 895 | All SZGR 2.0 genes in this pathway |
| KIM ALL DISORDERS CALB1 CORR UP | 548 | 370 | All SZGR 2.0 genes in this pathway |
| PURBEY TARGETS OF CTBP1 AND SATB1 DN | 180 | 116 | All SZGR 2.0 genes in this pathway |
| BOSCO ALLERGEN INDUCED TH2 ASSOCIATED MODULE | 151 | 86 | All SZGR 2.0 genes in this pathway |