Gene Page: NLGN1
Summary ?
| GeneID | 22871 |
| Symbol | NLGN1 |
| Synonyms | NL1 |
| Description | neuroligin 1 |
| Reference | MIM:600568|HGNC:HGNC:14291|Ensembl:ENSG00000169760|HPRD:07195|Vega:OTTHUMG00000157005 |
| Gene type | protein-coding |
| Map location | 3q26.31 |
| Pascal p-value | 2.755E-4 |
| Sherlock p-value | 0.004 |
| Fetal beta | 0.691 |
| Support | CELL ADHESION AND TRANSSYNAPTIC SIGNALING G2Cdb.humanPSD G2Cdb.humanPSP Potential synaptic genes |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| CV:PheWAS | Phenome-wide association studies (PheWAS) | 157 SNPs associated with schizophrenia | 1 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 5 |
Section I. Genetics and epigenetics annotation
 CV:PheWAS
CV:PheWAS
| SNP ID | Chromosome | Position | Allele | P | Function | Gene | Up/Down Distance |
|---|---|---|---|---|---|---|---|
| rs13074924 | 3 | 174029045 | null | 1.615 | NLGN1 | null |
Section II. Transcriptome annotation
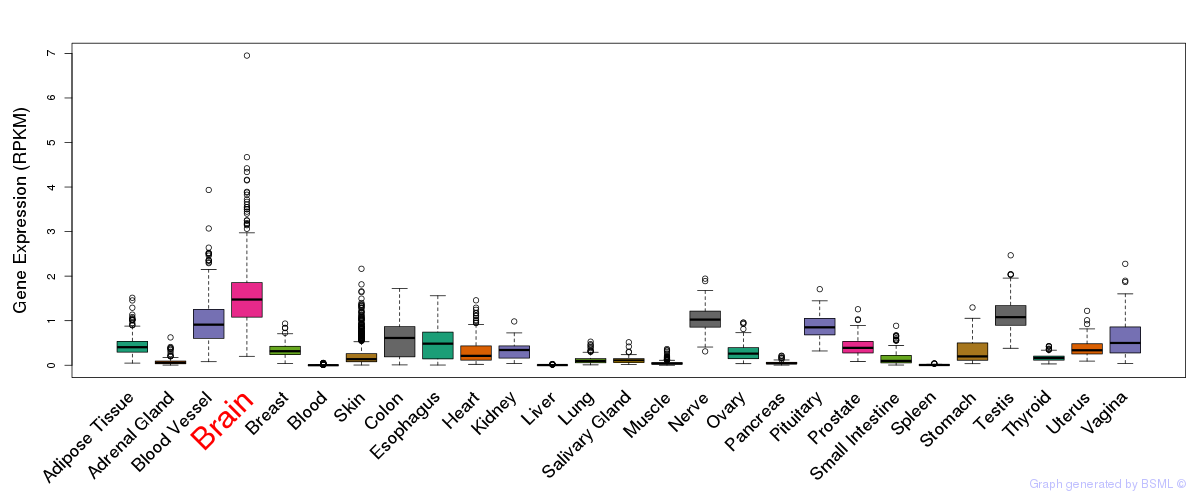
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| NCOA7 | 0.95 | 0.90 |
| COL4A3BP | 0.94 | 0.92 |
| CHM | 0.94 | 0.91 |
| PREPL | 0.94 | 0.88 |
| ARFGEF2 | 0.93 | 0.90 |
| MAP2K4 | 0.93 | 0.88 |
| PIK3CB | 0.93 | 0.90 |
| DPP8 | 0.93 | 0.87 |
| ATRNL1 | 0.93 | 0.90 |
| ATP6V1A | 0.92 | 0.86 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| C1orf61 | -0.50 | -0.64 |
| RAB34 | -0.49 | -0.55 |
| AF347015.21 | -0.48 | -0.46 |
| EFEMP2 | -0.47 | -0.53 |
| PLA2G5 | -0.47 | -0.46 |
| IL32 | -0.47 | -0.50 |
| DBI | -0.47 | -0.55 |
| RHOC | -0.47 | -0.58 |
| GATSL3 | -0.46 | -0.53 |
| ENHO | -0.46 | -0.54 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0042043 | neurexin binding | ISS | Synap (GO term level: 4) | - |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007416 | synaptogenesis | ISS | Synap (GO term level: 6) | - |
| GO:0016080 | synaptic vesicle targeting | ISS | Synap (GO term level: 10) | - |
| GO:0045664 | regulation of neuron differentiation | ISS | neuron, neurogenesis (GO term level: 9) | - |
| GO:0007155 | cell adhesion | IEA | - | |
| GO:0006605 | protein targeting | ISS | - | |
| GO:0016339 | calcium-dependent cell-cell adhesion | ISS | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0045211 | postsynaptic membrane | IEA | Synap, Neurotransmitter (GO term level: 5) | - |
| GO:0045202 | synapse | ISS | neuron, Synap, Neurotransmitter, Glial (GO term level: 2) | - |
| GO:0005886 | plasma membrane | IEA | - | |
| GO:0005887 | integral to plasma membrane | ISS | - | |
| GO:0030054 | cell junction | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG CELL ADHESION MOLECULES CAMS | 134 | 93 | All SZGR 2.0 genes in this pathway |
| ROVERSI GLIOMA COPY NUMBER UP | 100 | 75 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 12HR UP | 116 | 79 | All SZGR 2.0 genes in this pathway |
| KAAB HEART ATRIUM VS VENTRICLE DN | 261 | 183 | All SZGR 2.0 genes in this pathway |
| BHATI G2M ARREST BY 2METHOXYESTRADIOL DN | 127 | 75 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS UP | 1305 | 895 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-15/16/195/424/497 | 645 | 651 | m8 | hsa-miR-15abrain | UAGCAGCACAUAAUGGUUUGUG |
| hsa-miR-16brain | UAGCAGCACGUAAAUAUUGGCG | ||||
| hsa-miR-15bbrain | UAGCAGCACAUCAUGGUUUACA | ||||
| hsa-miR-195SZ | UAGCAGCACAGAAAUAUUGGC | ||||
| hsa-miR-424 | CAGCAGCAAUUCAUGUUUUGAA | ||||
| hsa-miR-497 | CAGCAGCACACUGUGGUUUGU | ||||
| miR-190 | 120 | 127 | 1A,m8 | hsa-miR-190 | UGAUAUGUUUGAUAUAUUAGGU |
| miR-193 | 508 | 514 | 1A | hsa-miR-193a | AACUGGCCUACAAAGUCCCAG |
| hsa-miR-193b | AACUGGCCCUCAAAGUCCCGCUUU | ||||
| miR-217 | 37 | 43 | 1A | hsa-miR-217 | UACUGCAUCAGGAACUGAUUGGAU |
| miR-30-5p | 1120 | 1126 | m8 | hsa-miR-30a-5p | UGUAAACAUCCUCGACUGGAAG |
| hsa-miR-30cbrain | UGUAAACAUCCUACACUCUCAGC | ||||
| hsa-miR-30dSZ | UGUAAACAUCCCCGACUGGAAG | ||||
| hsa-miR-30bSZ | UGUAAACAUCCUACACUCAGCU | ||||
| hsa-miR-30e-5p | UGUAAACAUCCUUGACUGGA | ||||
| hsa-miR-30a-5p | UGUAAACAUCCUCGACUGGAAG | ||||
| hsa-miR-30cbrain | UGUAAACAUCCUACACUCUCAGC | ||||
| hsa-miR-30dSZ | UGUAAACAUCCCCGACUGGAAG | ||||
| hsa-miR-30bSZ | UGUAAACAUCCUACACUCAGCU | ||||
| hsa-miR-30e-5p | UGUAAACAUCCUUGACUGGA | ||||
| miR-369-3p | 1640 | 1646 | 1A | hsa-miR-369-3p | AAUAAUACAUGGUUGAUCUUU |
| miR-374 | 1640 | 1646 | m8 | hsa-miR-374 | UUAUAAUACAACCUGAUAAGUG |
| miR-381 | 1654 | 1660 | 1A | hsa-miR-381 | UAUACAAGGGCAAGCUCUCUGU |
| hsa-miR-381 | UAUACAAGGGCAAGCUCUCUGU | ||||
| miR-448 | 354 | 360 | m8 | hsa-miR-448 | UUGCAUAUGUAGGAUGUCCCAU |
| miR-493-5p | 1399 | 1405 | m8 | hsa-miR-493-5p | UUGUACAUGGUAGGCUUUCAUU |
| miR-495 | 971 | 977 | 1A | hsa-miR-495brain | AAACAAACAUGGUGCACUUCUUU |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.