Gene Page: FOXG1
Summary ?
| GeneID | 2290 |
| Symbol | FOXG1 |
| Synonyms | BF1|BF2|FHKL3|FKH2|FKHL1|FKHL2|FKHL3|FKHL4|FOXG1A|FOXG1B|FOXG1C|HBF-1|HBF-2|HBF-3|HBF-G2|HBF2|HFK1|HFK2|HFK3|KHL2|QIN |
| Description | forkhead box G1 |
| Reference | MIM:164874|HGNC:HGNC:3811|HPRD:01283| |
| Gene type | protein-coding |
| Map location | 14q13 |
| Pascal p-value | 0.167 |
| Sherlock p-value | 0.84 |
| Fetal beta | 2.6 |
| DMG | 1 (# studies) |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 1 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| GSMA_I | Genome scan meta-analysis | Psr: 0.047 | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 5 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg21923619 | 14 | 29235469 | FOXG1 | 9.88E-9 | -0.029 | 4.33E-6 | DMG:Jaffe_2016 |
Section II. Transcriptome annotation
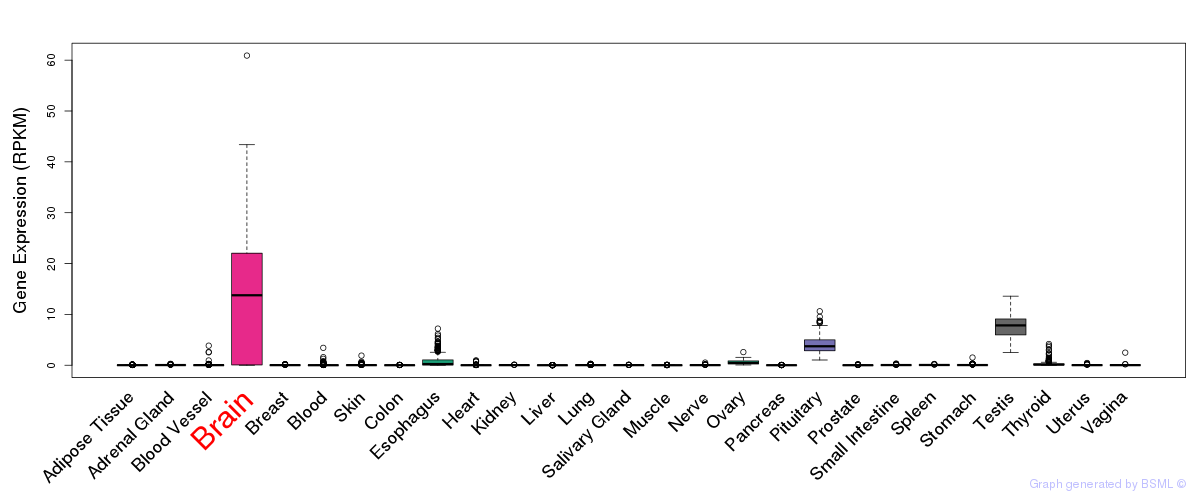
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| FOXC2 | 0.85 | 0.62 |
| CFH | 0.79 | 0.58 |
| GJB2 | 0.78 | 0.49 |
| SIX2 | 0.78 | 0.42 |
| OGN | 0.76 | 0.47 |
| SCARA5 | 0.75 | 0.40 |
| DSP | 0.75 | 0.47 |
| SIX1 | 0.74 | 0.46 |
| ITGBL1 | 0.74 | 0.42 |
| OSR1 | 0.71 | 0.38 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| C1orf63 | -0.17 | -0.17 |
| AC007216.4 | -0.16 | -0.15 |
| SLC25A41 | -0.15 | -0.14 |
| SNHG12 | -0.15 | -0.31 |
| C11orf51 | -0.15 | -0.21 |
| CRYGS | -0.15 | -0.10 |
| KCNMB3 | -0.14 | -0.24 |
| CYP2C8 | -0.14 | -0.18 |
| NPIP | -0.14 | -0.10 |
| DFNB59 | -0.14 | -0.12 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0003700 | transcription factor activity | TAS | - | |
| GO:0043565 | sequence-specific DNA binding | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0048667 | cell morphogenesis involved in neuron differentiation | IEA | neuron (GO term level: 10) | - |
| GO:0016199 | axon midline choice point recognition | IEA | axon (GO term level: 15) | - |
| GO:0030900 | forebrain development | IEA | Brain (GO term level: 8) | - |
| GO:0021954 | central nervous system neuron development | IEA | neuron (GO term level: 10) | - |
| GO:0045665 | negative regulation of neuron differentiation | IEA | neuron (GO term level: 10) | - |
| GO:0002052 | positive regulation of neuroblast proliferation | IEA | - | |
| GO:0006355 | regulation of transcription, DNA-dependent | NAS | - | |
| GO:0006350 | transcription | IEA | - | |
| GO:0007346 | regulation of mitotic cell cycle | IEA | - | |
| GO:0009953 | dorsal/ventral pattern formation | IEA | - | |
| GO:0007275 | multicellular organismal development | IEA | - | |
| GO:0042472 | inner ear morphogenesis | IEA | - | |
| GO:0045787 | positive regulation of cell cycle | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005634 | nucleus | NAS | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| EEF1G | EF1G | GIG35 | eukaryotic translation elongation factor 1 gamma | Two-hybrid | BioGRID | 16169070 |
| FOXH1 | FAST-1 | FAST1 | forkhead box H1 | - | HPRD,BioGRID | 10938097 |
| FOXO1 | FKH1 | FKHR | FOXO1A | forkhead box O1 | FoxO1 interacts with FoxG1. This interaction was modelled on an interaction demonstrated between FoxO1 from an unspecified species and mouse FoxG1. | BIND | 15084259 |
| FOXO3 | AF6q21 | DKFZp781A0677 | FKHRL1 | FKHRL1P2 | FOXO2 | FOXO3A | MGC12739 | MGC31925 | forkhead box O3 | FoxO3 interacts with FoxG1. | BIND | 15084259 |
| FOXO4 | AFX | AFX1 | MGC120490 | MLLT7 | forkhead box O4 | FoxO4 interacts with FoxG1. This interaction was modelled on a demonstrated interaction between human FoxO4 and mouse FoxG1. | BIND | 15084259 |
| GDF9 | - | growth differentiation factor 9 | Two-hybrid | BioGRID | 16169070 |
| HDAC1 | DKFZp686H12203 | GON-10 | HD1 | RPD3 | RPD3L1 | histone deacetylase 1 | - | HPRD | 11238932 |
| HES1 | FLJ20408 | HES-1 | HHL | HRY | bHLHb39 | hairy and enhancer of split 1, (Drosophila) | - | HPRD | 11238932 |
| JARID1B | FLJ10538 | FLJ12459 | FLJ12491 | FLJ16281 | FLJ23670 | KDM5B | PLU-1 | PUT1 | RBBP2H1A | jumonji, AT rich interactive domain 1B | - | HPRD,BioGRID | 12657635 |
| SMAD1 | BSP1 | JV4-1 | JV41 | MADH1 | MADR1 | SMAD family member 1 | - | HPRD,BioGRID | 11387330 |
| TLE1 | ESG | ESG1 | GRG1 | transducin-like enhancer of split 1 (E(sp1) homolog, Drosophila) | - | HPRD | 11238932 |
| TLE3 | ESG | ESG3 | FLJ39460 | GRG3 | HsT18976 | KIAA1547 | transducin-like enhancer of split 3 (E(sp1) homolog, Drosophila) | - | HPRD | 11238932 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| PID SMAD2 3NUCLEAR PATHWAY | 82 | 63 | All SZGR 2.0 genes in this pathway |
| LEE NEURAL CREST STEM CELL DN | 118 | 79 | All SZGR 2.0 genes in this pathway |
| LI CISPLATIN RESISTANCE DN | 35 | 20 | All SZGR 2.0 genes in this pathway |
| MARTORIATI MDM4 TARGETS NEUROEPITHELIUM DN | 164 | 111 | All SZGR 2.0 genes in this pathway |
| DAWSON METHYLATED IN LYMPHOMA TCL1 | 59 | 45 | All SZGR 2.0 genes in this pathway |
| GROSS HYPOXIA VIA HIF1A DN | 110 | 78 | All SZGR 2.0 genes in this pathway |
| BENPORATH SUZ12 TARGETS | 1038 | 678 | All SZGR 2.0 genes in this pathway |
| BENPORATH EED TARGETS | 1062 | 725 | All SZGR 2.0 genes in this pathway |
| BENPORATH ES WITH H3K27ME3 | 1118 | 744 | All SZGR 2.0 genes in this pathway |
| BENPORATH PRC2 TARGETS | 652 | 441 | All SZGR 2.0 genes in this pathway |
| BENPORATH ES CORE NINE CORRELATED | 100 | 68 | All SZGR 2.0 genes in this pathway |
| BYSTRYKH HEMATOPOIESIS STEM CELL QTL TRANS | 882 | 572 | All SZGR 2.0 genes in this pathway |
| GERY CEBP TARGETS | 126 | 90 | All SZGR 2.0 genes in this pathway |
| LU AGING BRAIN DN | 153 | 120 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE DN | 1237 | 837 | All SZGR 2.0 genes in this pathway |
| PAL PRMT5 TARGETS UP | 203 | 135 | All SZGR 2.0 genes in this pathway |
| WANG LSD1 TARGETS UP | 24 | 14 | All SZGR 2.0 genes in this pathway |
| HELLER HDAC TARGETS SILENCED BY METHYLATION UP | 461 | 298 | All SZGR 2.0 genes in this pathway |
| BLUM RESPONSE TO SALIRASIB DN | 342 | 220 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS UP | 1305 | 895 | All SZGR 2.0 genes in this pathway |
| SCHRAETS MLL TARGETS UP | 35 | 21 | All SZGR 2.0 genes in this pathway |
| LI WILMS TUMOR | 27 | 12 | All SZGR 2.0 genes in this pathway |
| SHEDDEN LUNG CANCER POOR SURVIVAL A6 | 456 | 285 | All SZGR 2.0 genes in this pathway |
| HAN SATB1 TARGETS UP | 395 | 249 | All SZGR 2.0 genes in this pathway |
| MEISSNER NPC HCP WITH H3K4ME2 AND H3K27ME3 | 349 | 234 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN NPC HCP WITH H3K27ME3 | 341 | 243 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN MEF HCP WITH H3K27ME3 | 590 | 403 | All SZGR 2.0 genes in this pathway |
| KIM ALL DISORDERS OLIGODENDROCYTE NUMBER CORR UP | 756 | 494 | All SZGR 2.0 genes in this pathway |
| KIM BIPOLAR DISORDER OLIGODENDROCYTE DENSITY CORR UP | 682 | 440 | All SZGR 2.0 genes in this pathway |
| KIM ALL DISORDERS CALB1 CORR UP | 548 | 370 | All SZGR 2.0 genes in this pathway |
| CARD MIR302A TARGETS | 77 | 62 | All SZGR 2.0 genes in this pathway |
| AZARE NEOPLASTIC TRANSFORMATION BY STAT3 DN | 17 | 10 | All SZGR 2.0 genes in this pathway |
| PLASARI TGFB1 SIGNALING VIA NFIC 1HR UP | 33 | 25 | All SZGR 2.0 genes in this pathway |
| PLASARI TGFB1 SIGNALING VIA NFIC 10HR DN | 30 | 25 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 2ND EGF PULSE ONLY | 1725 | 838 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-133 | 8 | 14 | m8 | hsa-miR-133a | UUGGUCCCCUUCAACCAGCUGU |
| hsa-miR-133b | UUGGUCCCCUUCAACCAGCUA | ||||
| miR-144 | 835 | 841 | m8 | hsa-miR-144 | UACAGUAUAGAUGAUGUACUAG |
| miR-186 | 477 | 483 | 1A | hsa-miR-186 | CAAAGAAUUCUCCUUUUGGGCUU |
| miR-200bc/429 | 315 | 322 | 1A,m8 | hsa-miR-200b | UAAUACUGCCUGGUAAUGAUGAC |
| hsa-miR-200c | UAAUACUGCCGGGUAAUGAUGG | ||||
| hsa-miR-429 | UAAUACUGUCUGGUAAAACCGU | ||||
| hsa-miR-200b | UAAUACUGCCUGGUAAUGAUGAC | ||||
| hsa-miR-200c | UAAUACUGCCGGGUAAUGAUGG | ||||
| hsa-miR-429 | UAAUACUGUCUGGUAAAACCGU | ||||
| miR-203.1 | 259 | 265 | 1A | hsa-miR-203 | UGAAAUGUUUAGGACCACUAG |
| miR-208 | 813 | 819 | 1A | hsa-miR-208 | AUAAGACGAGCAAAAAGCUUGU |
| miR-217 | 313 | 319 | 1A | hsa-miR-217 | UACUGCAUCAGGAACUGAUUGGAU |
| miR-25/32/92/363/367 | 172 | 178 | m8 | hsa-miR-25brain | CAUUGCACUUGUCUCGGUCUGA |
| hsa-miR-32 | UAUUGCACAUUACUAAGUUGC | ||||
| hsa-miR-92 | UAUUGCACUUGUCCCGGCCUG | ||||
| hsa-miR-367 | AAUUGCACUUUAGCAAUGGUGA | ||||
| hsa-miR-92bSZ | UAUUGCACUCGUCCCGGCCUC | ||||
| miR-299-5p | 746 | 752 | m8 | hsa-miR-299-5p | UGGUUUACCGUCCCACAUACAU |
| miR-30-5p | 874 | 880 | m8 | hsa-miR-30a-5p | UGUAAACAUCCUCGACUGGAAG |
| hsa-miR-30cbrain | UGUAAACAUCCUACACUCUCAGC | ||||
| hsa-miR-30dSZ | UGUAAACAUCCCCGACUGGAAG | ||||
| hsa-miR-30bSZ | UGUAAACAUCCUACACUCAGCU | ||||
| hsa-miR-30e-5p | UGUAAACAUCCUUGACUGGA | ||||
| hsa-miR-30a-5p | UGUAAACAUCCUCGACUGGAAG | ||||
| hsa-miR-30cbrain | UGUAAACAUCCUACACUCUCAGC | ||||
| hsa-miR-30dSZ | UGUAAACAUCCCCGACUGGAAG | ||||
| hsa-miR-30bSZ | UGUAAACAUCCUACACUCAGCU | ||||
| hsa-miR-30e-5p | UGUAAACAUCCUUGACUGGA | ||||
| miR-330 | 251 | 257 | m8 | hsa-miR-330brain | GCAAAGCACACGGCCUGCAGAGA |
| miR-34/449 | 599 | 605 | m8 | hsa-miR-34abrain | UGGCAGUGUCUUAGCUGGUUGUU |
| hsa-miR-34c | AGGCAGUGUAGUUAGCUGAUUGC | ||||
| hsa-miR-449 | UGGCAGUGUAUUGUUAGCUGGU | ||||
| hsa-miR-449b | AGGCAGUGUAUUGUUAGCUGGC | ||||
| miR-369-3p | 317 | 324 | 1A,m8 | hsa-miR-369-3p | AAUAAUACAUGGUUGAUCUUU |
| miR-374 | 318 | 325 | 1A,m8 | hsa-miR-374 | UUAUAAUACAACCUGAUAAGUG |
| miR-376 | 880 | 886 | 1A | hsa-miR-376a | AUCAUAGAGGAAAAUCCACGU |
| hsa-miR-376b | AUCAUAGAGGAAAAUCCAUGUU | ||||
| miR-378* | 70 | 76 | m8 | hsa-miR-422b | CUGGACUUGGAGUCAGAAGGCC |
| hsa-miR-422a | CUGGACUUAGGGUCAGAAGGCC | ||||
| miR-499 | 813 | 819 | 1A | hsa-miR-499 | UUAAGACUUGCAGUGAUGUUUAA |
| miR-500 | 170 | 177 | 1A,m8 | hsa-miR-500 | AUGCACCUGGGCAAGGAUUCUG |
| miR-543 | 392 | 398 | 1A | hsa-miR-543 | AAACAUUCGCGGUGCACUUCU |
| hsa-miR-543 | AAACAUUCGCGGUGCACUUCU | ||||
| miR-9 | 518 | 525 | 1A,m8 | hsa-miR-9SZ | UCUUUGGUUAUCUAGCUGUAUGA |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.