Gene Page: KDM2A
Summary ?
| GeneID | 22992 |
| Symbol | KDM2A |
| Synonyms | CXXC8|FBL11|FBL7|FBXL11|JHDM1A|LILINA |
| Description | lysine demethylase 2A |
| Reference | MIM:605657|HGNC:HGNC:13606|Ensembl:ENSG00000173120|HPRD:05741|Vega:OTTHUMG00000167103 |
| Gene type | protein-coding |
| Map location | 11q13.2 |
| Pascal p-value | 0.386 |
| Sherlock p-value | 0.369 |
| Fetal beta | 0.155 |
| DMG | 1 (# studies) |
| eGene | Myers' cis & trans |
| Support | Chromatin Remodeling Genes |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg21293464 | 11 | 66885276 | KDM2A | 2.082E-4 | 0.288 | 0.035 | DMG:Wockner_2014 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs16829545 | chr2 | 151977407 | KDM2A | 22992 | 0.01 | trans | ||
| rs16955618 | chr15 | 29937543 | KDM2A | 22992 | 0.03 | trans | ||
| rs17264923 | chrX | 69499164 | KDM2A | 22992 | 0.12 | trans |
Section II. Transcriptome annotation
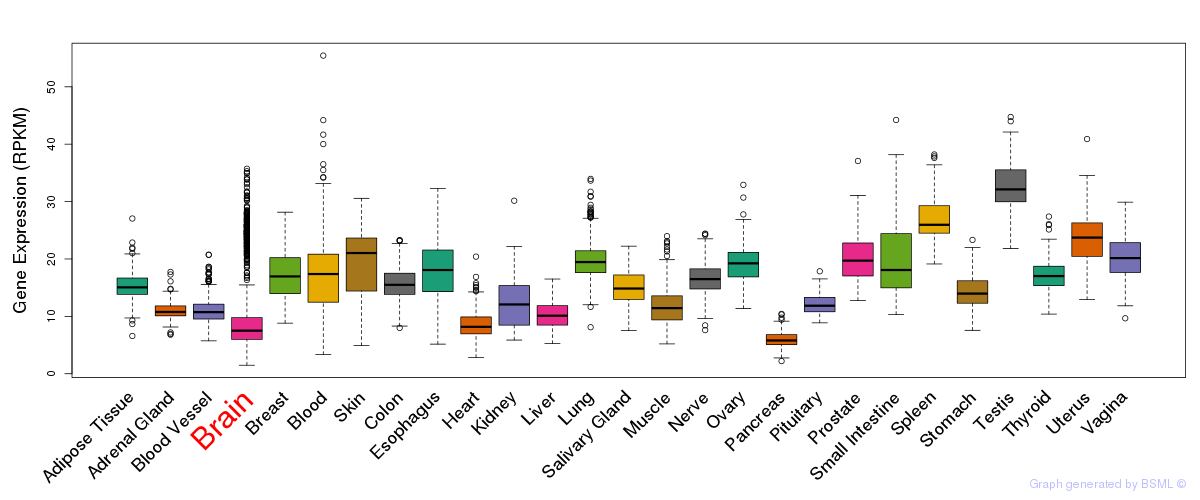
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| EGR2 | 0.89 | 0.81 |
| CX3CL1 | 0.82 | 0.83 |
| EGR1 | 0.80 | 0.81 |
| GPR26 | 0.79 | 0.68 |
| FMNL1 | 0.79 | 0.84 |
| TMEM132D | 0.77 | 0.80 |
| COL10A1 | 0.77 | 0.61 |
| GRIN2A | 0.76 | 0.78 |
| NR4A1 | 0.76 | 0.74 |
| CTSB | 0.76 | 0.80 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| HEBP2 | -0.39 | -0.69 |
| GPR125 | -0.38 | -0.42 |
| UBE2L6 | -0.37 | -0.38 |
| BCL7C | -0.36 | -0.53 |
| FADS2 | -0.36 | -0.39 |
| RPL23A | -0.36 | -0.53 |
| TUBB2B | -0.36 | -0.46 |
| TRAF4 | -0.36 | -0.46 |
| KIAA1949 | -0.35 | -0.35 |
| DYNLT1 | -0.35 | -0.55 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| DEURIG T CELL PROLYMPHOCYTIC LEUKEMIA DN | 320 | 184 | All SZGR 2.0 genes in this pathway |
| HORIUCHI WTAP TARGETS UP | 306 | 188 | All SZGR 2.0 genes in this pathway |
| HAHTOLA SEZARY SYNDROM DN | 42 | 26 | All SZGR 2.0 genes in this pathway |
| MISSIAGLIA REGULATED BY METHYLATION UP | 126 | 78 | All SZGR 2.0 genes in this pathway |
| PEREZ TP53 TARGETS | 1174 | 695 | All SZGR 2.0 genes in this pathway |
| PATTERSON DOCETAXEL RESISTANCE | 29 | 20 | All SZGR 2.0 genes in this pathway |
| AMUNDSON RESPONSE TO ARSENITE | 217 | 143 | All SZGR 2.0 genes in this pathway |
| BUYTAERT PHOTODYNAMIC THERAPY STRESS UP | 811 | 508 | All SZGR 2.0 genes in this pathway |
| LOCKWOOD AMPLIFIED IN LUNG CANCER | 214 | 139 | All SZGR 2.0 genes in this pathway |
| BENPORATH NANOG TARGETS | 988 | 594 | All SZGR 2.0 genes in this pathway |
| ZHANG RESPONSE TO IKK INHIBITOR AND TNF UP | 223 | 140 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| DAZARD RESPONSE TO UV SCC DN | 123 | 86 | All SZGR 2.0 genes in this pathway |
| CAIRO HEPATOBLASTOMA CLASSES DN | 210 | 141 | All SZGR 2.0 genes in this pathway |
| KRIEG HYPOXIA NOT VIA KDM3A | 770 | 480 | All SZGR 2.0 genes in this pathway |
| PHONG TNF RESPONSE VIA P38 COMPLETE | 227 | 151 | All SZGR 2.0 genes in this pathway |