Gene Page: KDM4C
Summary ?
| GeneID | 23081 |
| Symbol | KDM4C |
| Synonyms | GASC1|JHDM3C|JMJD2C|TDRD14C |
| Description | lysine demethylase 4C |
| Reference | MIM:605469|HGNC:HGNC:17071|Ensembl:ENSG00000107077|Ensembl:ENSG00000274527|Ensembl:ENSG00000282960|HPRD:12016|Vega:OTTHUMG00000019536Vega:OTTHUMG00000187508 |
| Gene type | protein-coding |
| Map location | 9p24.1 |
| Pascal p-value | 1.313E-4 |
| Fetal beta | -0.567 |
| DMG | 1 (# studies) |
| eGene | Cerebellar Hemisphere Cerebellum |
| Support | Chromatin Remodeling Genes |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWAScat | Genome-wide Association Studies | This data set includes 560 SNPs associated with schizophrenia. A total of 486 genes were mapped to these SNPs within 50kb. | |
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg14473811 | 9 | 6757912 | KDM4C | 2.73E-9 | -0.009 | 1.95E-6 | DMG:Jaffe_2016 |
Section II. Transcriptome annotation
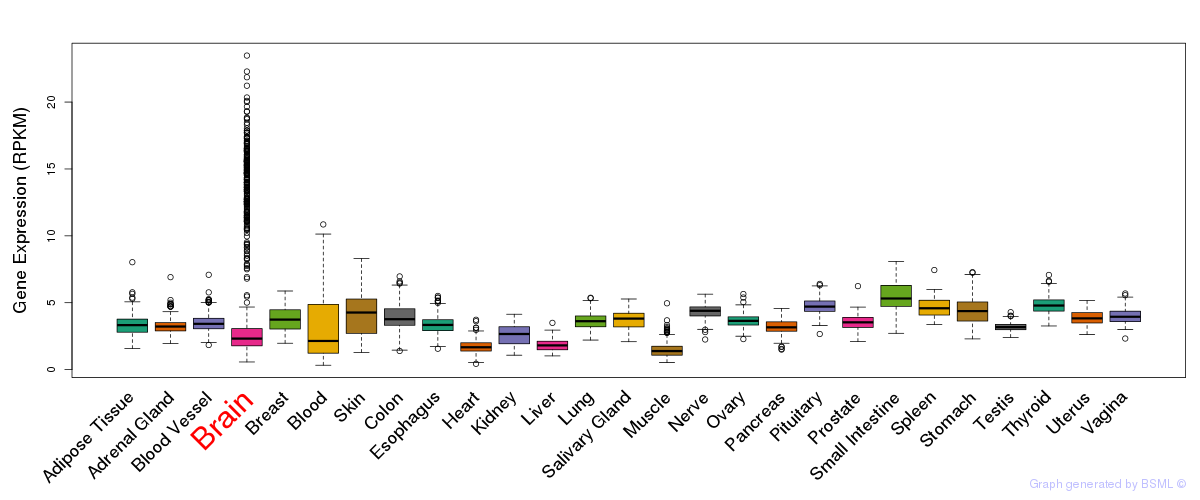
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| TGFBRAP1 | 0.96 | 0.97 |
| ZNF398 | 0.96 | 0.96 |
| URB1 | 0.96 | 0.97 |
| DIP2C | 0.96 | 0.96 |
| FOXK2 | 0.95 | 0.96 |
| SEC16A | 0.95 | 0.96 |
| ZNF317 | 0.95 | 0.96 |
| MTMR4 | 0.95 | 0.97 |
| LDOC1L | 0.95 | 0.97 |
| SEC24C | 0.95 | 0.96 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.31 | -0.76 | -0.88 |
| MT-CO2 | -0.75 | -0.88 |
| AF347015.27 | -0.73 | -0.85 |
| FXYD1 | -0.73 | -0.84 |
| AF347015.33 | -0.72 | -0.82 |
| MT-CYB | -0.72 | -0.83 |
| AF347015.8 | -0.72 | -0.86 |
| HIGD1B | -0.72 | -0.85 |
| S100B | -0.71 | -0.81 |
| IFI27 | -0.70 | -0.81 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| PID AR PATHWAY | 61 | 46 | All SZGR 2.0 genes in this pathway |
| DAVICIONI MOLECULAR ARMS VS ERMS UP | 332 | 228 | All SZGR 2.0 genes in this pathway |
| RODRIGUES NTN1 TARGETS DN | 158 | 102 | All SZGR 2.0 genes in this pathway |
| PROVENZANI METASTASIS UP | 194 | 112 | All SZGR 2.0 genes in this pathway |
| SNIJDERS AMPLIFIED IN HEAD AND NECK TUMORS | 37 | 27 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 DN | 855 | 609 | All SZGR 2.0 genes in this pathway |
| NUYTTEN NIPP1 TARGETS UP | 769 | 437 | All SZGR 2.0 genes in this pathway |
| NUYTTEN EZH2 TARGETS UP | 1037 | 673 | All SZGR 2.0 genes in this pathway |
| PARK HSC VS MULTIPOTENT PROGENITORS DN | 18 | 12 | All SZGR 2.0 genes in this pathway |
| MENSE HYPOXIA UP | 98 | 71 | All SZGR 2.0 genes in this pathway |
| SU SALIVARY GLAND | 16 | 9 | All SZGR 2.0 genes in this pathway |
| CUI TCF21 TARGETS 2 DN | 830 | 547 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 48HR UP | 180 | 125 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 18HR UP | 178 | 111 | All SZGR 2.0 genes in this pathway |