Gene Page: ZC3H4
Summary ?
| GeneID | 23211 |
| Symbol | ZC3H4 |
| Synonyms | C19orf7 |
| Description | zinc finger CCCH-type containing 4 |
| Reference | HGNC:HGNC:17808|Ensembl:ENSG00000130749|Vega:OTTHUMG00000183442 |
| Gene type | protein-coding |
| Map location | 19q13.32 |
| Pascal p-value | 0.095 |
| Fetal beta | 0.988 |
| DMG | 2 (# studies) |
| eGene | Nucleus accumbens basal ganglia Meta |
| Support | CompositeSet Darnell FMRP targets Ascano FMRP targets |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Montano_2016 | Genome-wide DNA methylation analysis | This dataset includes 172 replicated associations between CpGs with schizophrenia. | 2 |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 2 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg27300045 | 19 | 47610813 | ZC3H4 | 3.93E-4 | -0.004 | 0.197 | DMG:Montano_2016 |
| cg18489475 | 19 | 47617233 | ZC3H4 | 5.296E-4 | 0.595 | 0.048 | DMG:Wockner_2014 |
Section II. Transcriptome annotation
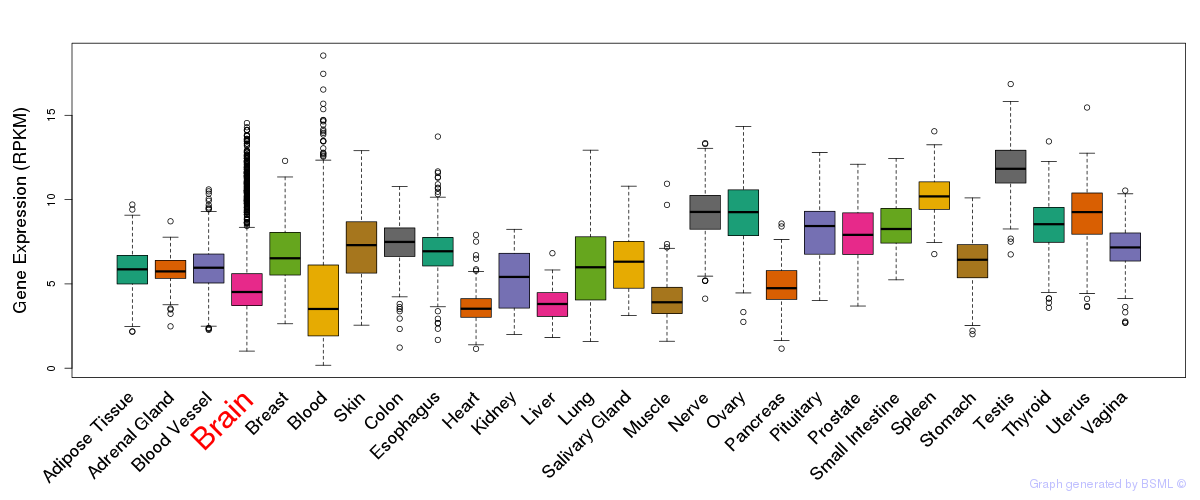
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| BRWD2 | 0.90 | 0.91 |
| WDR5 | 0.88 | 0.88 |
| CUL4A | 0.87 | 0.89 |
| KDM2A | 0.87 | 0.89 |
| MBTPS1 | 0.87 | 0.88 |
| SFRS14 | 0.87 | 0.87 |
| POLS | 0.87 | 0.89 |
| SEC23IP | 0.87 | 0.88 |
| GNE | 0.87 | 0.88 |
| MCM3AP | 0.87 | 0.87 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| MT-CO2 | -0.77 | -0.80 |
| AF347015.31 | -0.77 | -0.80 |
| AF347015.21 | -0.75 | -0.83 |
| FXYD1 | -0.73 | -0.75 |
| AF347015.27 | -0.73 | -0.75 |
| MT-CYB | -0.73 | -0.75 |
| AF347015.8 | -0.73 | -0.77 |
| AF347015.33 | -0.73 | -0.74 |
| HIGD1B | -0.73 | -0.78 |
| IFI27 | -0.71 | -0.72 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| DACOSTA UV RESPONSE VIA ERCC3 DN | 855 | 609 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 COMMON DN | 483 | 336 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA1 PCC NETWORK | 1652 | 1023 | All SZGR 2.0 genes in this pathway |
| PUJANA ATM PCC NETWORK | 1442 | 892 | All SZGR 2.0 genes in this pathway |
| NOUZOVA TRETINOIN AND H4 ACETYLATION | 143 | 85 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| GENTILE UV RESPONSE CLUSTER D2 | 41 | 30 | All SZGR 2.0 genes in this pathway |
| GENTILE UV HIGH DOSE DN | 312 | 203 | All SZGR 2.0 genes in this pathway |
| DAZARD RESPONSE TO UV NHEK DN | 318 | 220 | All SZGR 2.0 genes in this pathway |
| MILI PSEUDOPODIA CHEMOTAXIS DN | 457 | 302 | All SZGR 2.0 genes in this pathway |
| MILI PSEUDOPODIA HAPTOTAXIS DN | 668 | 419 | All SZGR 2.0 genes in this pathway |
| MARTENS BOUND BY PML RARA FUSION | 456 | 287 | All SZGR 2.0 genes in this pathway |
| JOHNSTONE PARVB TARGETS 3 DN | 918 | 550 | All SZGR 2.0 genes in this pathway |
| GOBERT OLIGODENDROCYTE DIFFERENTIATION UP | 570 | 339 | All SZGR 2.0 genes in this pathway |