Gene Page: PPIP5K2
Summary ?
| GeneID | 23262 |
| Symbol | PPIP5K2 |
| Synonyms | CFAP160|HISPPD1|IP7K2|VIP2 |
| Description | diphosphoinositol pentakisphosphate kinase 2 |
| Reference | MIM:611648|HGNC:HGNC:29035|Ensembl:ENSG00000145725|HPRD:13802|Vega:OTTHUMG00000181461 |
| Gene type | protein-coding |
| Map location | 5q21.1 |
| Pascal p-value | 0.349 |
| DMG | 1 (# studies) |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg08295258 | 5 | 102455984 | PPIP5K2 | 3.73E-8 | -0.008 | 1.08E-5 | DMG:Jaffe_2016 |
Section II. Transcriptome annotation
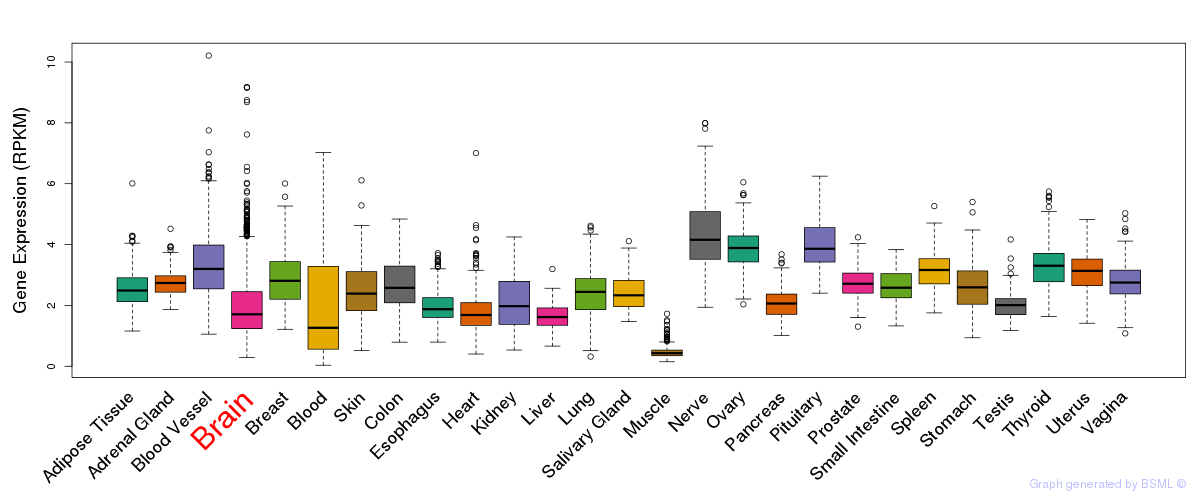
General gene expression (GTEx)
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AP000751.3 | 0.92 | 0.91 |
| SLC39A10 | 0.91 | 0.88 |
| CLOCK | 0.91 | 0.87 |
| UBR3 | 0.90 | 0.87 |
| ARFGEF2 | 0.90 | 0.88 |
| PLCB1 | 0.90 | 0.87 |
| CNKSR2 | 0.90 | 0.91 |
| BSN | 0.90 | 0.86 |
| ATRNL1 | 0.90 | 0.87 |
| KCNB1 | 0.89 | 0.87 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| RAB34 | -0.48 | -0.57 |
| DBI | -0.47 | -0.57 |
| RHOC | -0.45 | -0.56 |
| EFEMP2 | -0.45 | -0.45 |
| C1orf61 | -0.44 | -0.61 |
| RAB13 | -0.43 | -0.52 |
| NUDT8 | -0.42 | -0.45 |
| NKAIN4 | -0.41 | -0.52 |
| RFXANK | -0.41 | -0.44 |
| C19orf36 | -0.41 | -0.48 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| ONKEN UVEAL MELANOMA UP | 783 | 507 | All SZGR 2.0 genes in this pathway |
| GRABARCZYK BCL11B TARGETS UP | 81 | 40 | All SZGR 2.0 genes in this pathway |
| HAMAI APOPTOSIS VIA TRAIL UP | 584 | 356 | All SZGR 2.0 genes in this pathway |
| SCHLOSSER SERUM RESPONSE DN | 712 | 443 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 DN | 855 | 609 | All SZGR 2.0 genes in this pathway |
| PUJANA ATM PCC NETWORK | 1442 | 892 | All SZGR 2.0 genes in this pathway |
| MOREAUX MULTIPLE MYELOMA BY TACI DN | 172 | 107 | All SZGR 2.0 genes in this pathway |
| CUI TCF21 TARGETS 2 DN | 830 | 547 | All SZGR 2.0 genes in this pathway |
| DAZARD RESPONSE TO UV NHEK DN | 318 | 220 | All SZGR 2.0 genes in this pathway |
| XU GH1 AUTOCRINE TARGETS DN | 142 | 94 | All SZGR 2.0 genes in this pathway |
| ZHANG BREAST CANCER PROGENITORS UP | 425 | 253 | All SZGR 2.0 genes in this pathway |
| RAO BOUND BY SALL4 ISOFORM B | 517 | 302 | All SZGR 2.0 genes in this pathway |